Figures & data
Figure 1. Genome circle map (A) and COG function annotation (B) of B. velezensis BA-26. (A) The circle map shows seven kinds of information from the outside to the inside: circle 1 is genome location information, circle 2 is GC content information, circle 3 is coding gene on positive chain (red label), circle 4 is coding gene on negative chain (green label), circle 5 is ncRNA information on the positive chain (blue mark), circle 6 is ncRNA information on the negative chain (purple label), and circle 7 marks the information of long fragment repeats (≥ 100 bp) in the genome (orange tagging). (B) Functional classification of COG genes of strain BA-26.
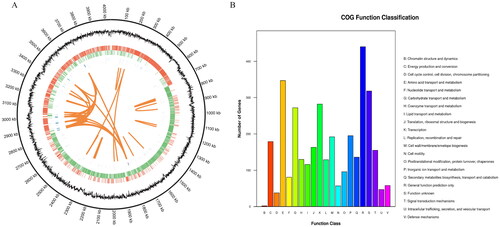
Table 1. Genomic characteristics of B. velezensis BA-26.
Supplemental Material
Download PDF (1.9 MB)Data availability
All data that support the findings from this study are available from the corresponding author (…) upon reasonable request.