Figures & data
Table 1. Best molecular docking results of selected compounds against the target proteins and their interactions.
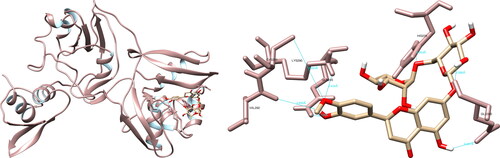
Figure 1. Genistein interactions with SARS-CoV-2 main protease, visualized in UCSF Chimera. Genistein formed six hydrogen bonds with amino acids: Cys44, Met49, Cys145, Glu166, Arg188 and Gln189.
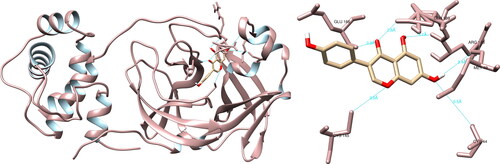
Figure 2. Hesperidin interactions with SARS-CoV-2 main protease, visualized in UCSF Chimera. Hesperidin formed seven hydrogen bonds with amino acids: Thr26, Asn142, Cys145, His164, Glu166 and Gln189.
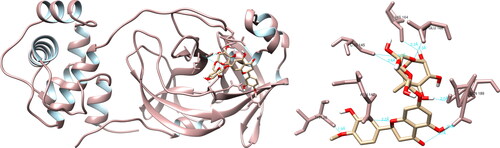
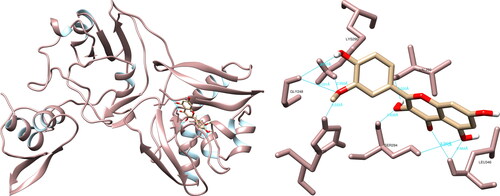
Figure 3. Hesperidin interactions with SARS-CoV-2 papain-like protease, visualized in UCSF Chimera. Hesperidin formed 11 hydrogen bonds with amino acids: Ile23, Asn37, Ala38, Asn40, Lys44, Gly48, Ala50, Gly51 and Ala129.
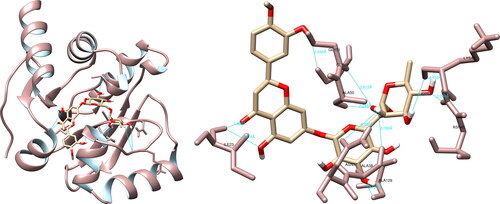
Figure 4. Luteolin interactions with SARS-CoV-2 papain-like protease, visualized in UCSF Chimera. Luteolin formed five hydrogen bonds with amino acids: His45, Gly46, Val49, Ala50 and Ser128.
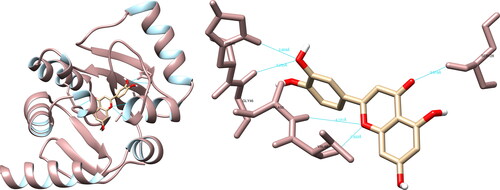
Figure 5. Hesperidin interactions with SARS-CoV-2 RNA-dependent RNA polymerase, visualized in UCSF Chimera. Hesperidin formed 10 hydrogen bonds with amino acids: Lys551, Tyr619, Arg624, Thr680, Asp760, Asp761 and Cys813.
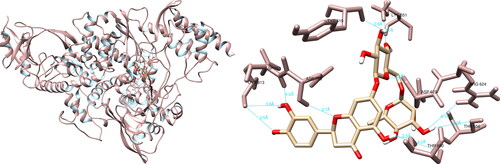
Figure 6. Quercetin interactions with SARS-CoV-2 RNA-dependent RNA polymerase, visualized in UCSF Chimera. Quercetin formed seven hydrogen bonds with amino acids: Arg553, Lys621, Cys622, Asp623 and Ser682.
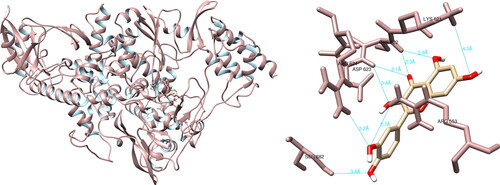
Figure 7. Hesperidin interactions with SARS-CoV-2 spike glycoprotein, visualized in UCSF Chimera. Hesperidin formed five hydrogen bonds with amino acids: Arg408, Phe490, Leu492, Gln498 and Tyr505.
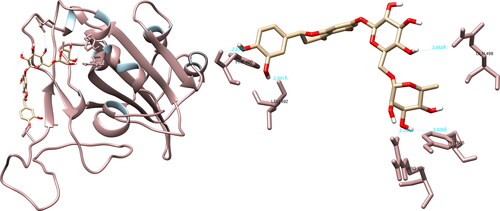
Figure 8. Fisetin interactions with SARS-CoV-2 spike glycoprotein, visualized in UCSF Chimera. Fisetin formed four hydrogen bonds with amino acids: Arg403, Gly496, Gln498 and Tyr505.
Supplemental Material
Download PDF (69.9 KB)Data availability
The datasets generated and/or analysed during the current study are available in the RCSB Protein Data Bank [https://www.rcsb.org/] and PubChem database repository [https://pubchem.ncbi.nlm.nih.gov/].
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.