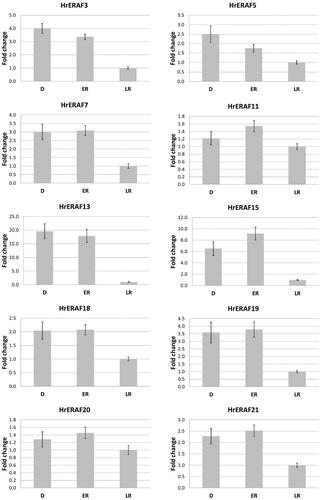
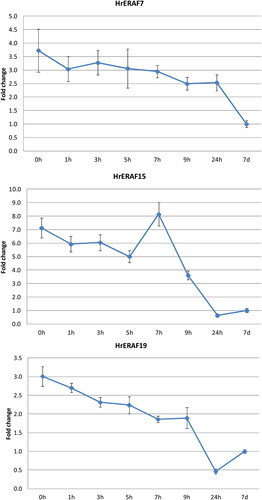
Figures & data
Figure 1. Representative example of oligo-dT anchored cDNA-SRAP analysis with three different SRAP primers (ME3, ME4 and ME5) during RAF: recovery after freezing-induced desiccation in H. rhodopensis. D: freezing desiccated state; ER: early recovery (1 + 3 h); LR: late recovery (24 h + 7 d); M: 100 bp Plus™; arrows indicate the differentially expressed bands that were selected for reamplification and sequencing.
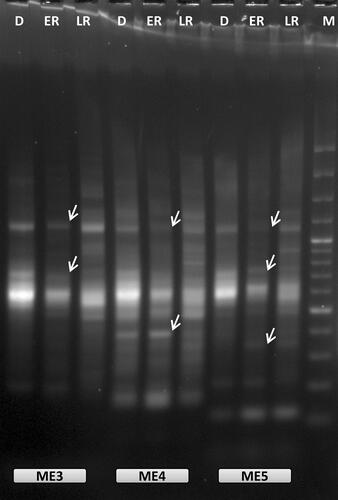
Figure 2. Examples of oligo-dT anchored cDNA-SCoT analysis with five different SCoT primers (SCoT1, SCoT2, SCoT8, SCoT9 and SCoT13) during RAF in H. rhodopensis. D: freezing desiccated state; ER: early recovery (1 + 3 h); LR: late recovery (24 h + 7 d); NTC: PCR reaction without cDNA; M: 100 bp Plus™; arrows indicate the differentially expressed bands that were selected for reamplification and sequencing.
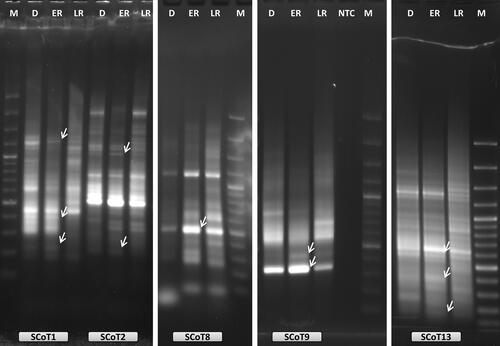
Table 1. Homology analysis of the selected differentially expressed TDFs to gene sequences in the NCBI non-redundant protein database using the BLASTX algorithm.