Figures & data
Table 1. Primers used for the amplification and sequencing of gene fragments from nonribosomal peptide synthetases.
Figure 1. Phylogenetic tree of Bacillus spp. based on 16S rRNA sequences through maximum-likelihood methods with 1000 bootstrap replications using MEGA version 6.0. Escherichia coli BL21(DE3) (AM946981.2) was used as an outgroup in the analysis. NCBI GenBank accession numbers used in the comparison reference strains are the following: B. velezensis FZB42, OR485707; B. amyloliquefaciens ATCC 23350, NR_118950; B. licheniformis ATCC 14580, NC_006270.3; B. aerius MN-1, MN252912; B. safensis ATCC BAA-1126T, AF234854; B. aryabhattai B8W22, NZ_JYOO01000023. NCBI acc. no. of the newly isolated strains are OR482395, OR482394, OR482396, OR482397, OP554433, MK461937, MK461933, MK461938, MK461934, MK461936, OR482398, OR482400, OR482399, MK461947.
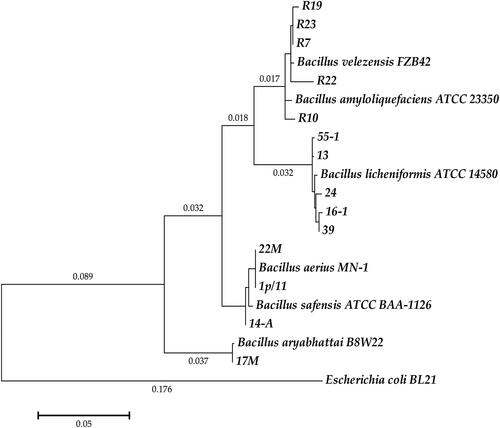
Table 2. Inhibition of Botrytis cinerea growth by cell-free supernatant of Bacillus spp. cultures obtained after 48 h of cultivation.
Table 3. Inhibition of Phytophthora infestans growth by the cell-free supernatants of 48-h Bacillus spp. cultures.
Table 4. Comparison of the antifungal activity against Phytophthora infestans displayed by B. velezensis R22 (BV R22) and B. licheniformis 39 (BL 39).
Figure 2. Antifungal activity of B. velezensis R22 against phytopathogenic fungi. (A) Alternaria alternata; (B) Aspergillus fumigatus; (C) Aspergillus niger; (D) Neocosmospora keratoplastica. The co-inoculation experiments were performed in 100 mm Petri dishes for 7 days.
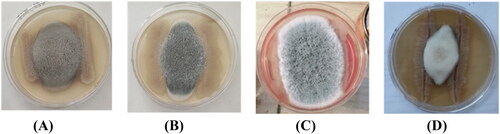
Table 5. Inhibition of R. solanacearum growth by cell-free supernatant of Bacillus spp. cultures obtained after 48 h of cultivation.
Figure 3. PCR amplification of fenA gene encoding fengycin synthetase A. Designations: M, molecular weight marker (Perfect PlusTM 1 kb DNA Ladder, EURx); 1, B. velezensis R7; 2, B. amyloliquefaciens R10; 3, B. velezensis R19; 4, B. velezensis R22; 5, B. velezensis R23; 6, negative control (E. coli DH5α).
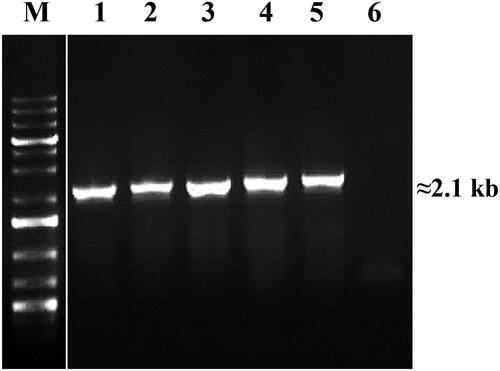
Figure 4. PCR amplification of specific fragments of srfAA gene encoding surfactin synthetase A (A) and lchAA gene for lichenysin synthase (B). Lanes and samples: M, molecular weight marker (Perfect PlusTM 1 kb DNA Ladder, EURx); (A): 1, B. velezensis R7; 2, B. amyloliquefaciens R10; 3, B. velezensis R19; 4, B. velezensis R22; 5, B. velezensis R23; 6, negative control (E. coli DH5α); (B) 1, B. licheniformis 16-1; 2, B. licheniformis 13; 3, B. licheniformis 24; 4, B. licheniformis 39; 5, B. licheniformis 55-1; 6, negative control (E. coli DH5α).
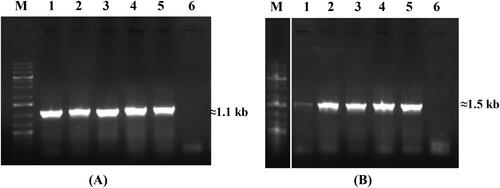
Table 6. Secondary metabolites predicted to be synthesized by B. velezensis R22 using AntiSMASH 7.0 platform.
Figure 5. Analyses of lipopeptides of B. velezensis strain R22 by UHPLC-Q-TOF mass spectrometer. (A) The total ion chromatogram of 10 μL lipopeptides extracted from the cell-free supenatant obtained after B. velezensis strain R22 growth, identified at λ 226 nm in the range m/z 150–3500; (B) LC-MS of a peak eluted between 14.7 and 16.3 min, predominant [M + H]+ ion at m/z 1036.698 corresponding to surfactin; (C) LC-MS of peak eluted between 4.4 and 5.9 min dominated by double charged ions [M + 2H]2+ at m/z 732.405, 739.412, 746.419, and 753.427 corresponding to [M + H]+ ions at m/z 1463.804, 1477.818, 1491.832, and 1505.849, identified as different forms of fengycin.
![Figure 5. Analyses of lipopeptides of B. velezensis strain R22 by UHPLC-Q-TOF mass spectrometer. (A) The total ion chromatogram of 10 μL lipopeptides extracted from the cell-free supenatant obtained after B. velezensis strain R22 growth, identified at λ 226 nm in the range m/z 150–3500; (B) LC-MS of a peak eluted between 14.7 and 16.3 min, predominant [M + H]+ ion at m/z 1036.698 corresponding to surfactin; (C) LC-MS of peak eluted between 4.4 and 5.9 min dominated by double charged ions [M + 2H]2+ at m/z 732.405, 739.412, 746.419, and 753.427 corresponding to [M + H]+ ions at m/z 1463.804, 1477.818, 1491.832, and 1505.849, identified as different forms of fengycin.](/cms/asset/b5bbfda5-ace9-4f0b-8615-8166c4d456bc/tbeq_a_2313072_f0005_b.jpg)
Figure 6. LC-MS/MS spectrum of the surfactin precursor [M + H]+ at m/z 1036.698, containing a C15 β-hydroxy fatty acid chain with interpretation.
![Figure 6. LC-MS/MS spectrum of the surfactin precursor [M + H]+ at m/z 1036.698, containing a C15 β-hydroxy fatty acid chain with interpretation.](/cms/asset/88ebd4c7-60fe-4579-80bf-3203d3851e1d/tbeq_a_2313072_f0006_b.jpg)
Figure 7. LC-MS/MS spectra of different forms of fengycin: (A) the precursor [M + 2H]2+ at m/z 732.405, containing Ala of position 6 and C16 β-hydroxy fatty acid chain; (B) the precursor [M + 2H]2+ at m/z 739.412, containing Ala of position 6 and C17 β-hydroxy fatty acid chain; (C) the precursor [M + 2H]2+ at m/z 746.419, containing Val of position 6 and C16 β-hydroxy fatty acid chain; (D) the precursor [M + 2H]2+ at m/z 753.427, containing Val of position 6 and C17 β-hydroxy fatty acid chain.
![Figure 7. LC-MS/MS spectra of different forms of fengycin: (A) the precursor [M + 2H]2+ at m/z 732.405, containing Ala of position 6 and C16 β-hydroxy fatty acid chain; (B) the precursor [M + 2H]2+ at m/z 739.412, containing Ala of position 6 and C17 β-hydroxy fatty acid chain; (C) the precursor [M + 2H]2+ at m/z 746.419, containing Val of position 6 and C16 β-hydroxy fatty acid chain; (D) the precursor [M + 2H]2+ at m/z 753.427, containing Val of position 6 and C17 β-hydroxy fatty acid chain.](/cms/asset/f964dd70-30cb-44dc-9581-7309c3ee01b8/tbeq_a_2313072_f0007_c.jpg)
Table 7. Lipopeptides established by mass spectrometry in cell-free fermentation broth (48-h culture) of B. velezensis strain R22.
Supplemental Material
Download PDF (1.1 MB)Data availability
All data concerning nucleotide sequences are available in the NCBI GenBank. De novo sequenced genome of B. velezensis strain R22 has been deposited in DDBJ/ENA/GenBank under accession no. JASUZW000000000.1, BioSample SAMN35683875. The version described in this article is the first version. The Sequence Read Archive is deposited under accession no SRP441980.
