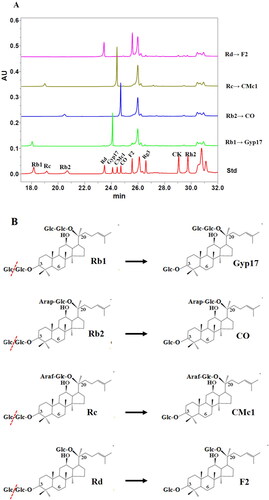
Figures & data
Figure 1. Isolation of endophytic fungi from Panax ginseng. Representative images. The purchased ginseng roots were washed and disinfected with 75% alcohol; then they were sliced and incubated for culture on a PDA medium supplemented with 100 mg/L ampicillin at 28 °C. (A) Screening endophytic fungi from ginseng roots. (B) Plate streaking to obtain single colonies. (C) Single colonies of endophytic fungi. (D) The rinse solution after the last disinfection of ginseng root as a negative control.
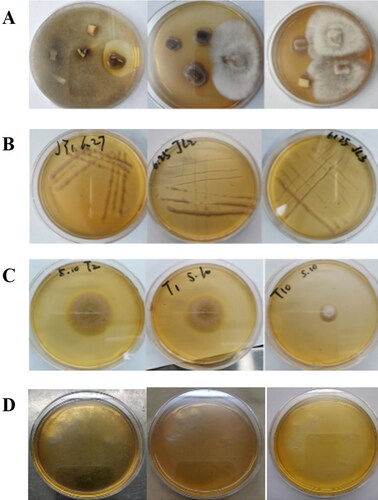
Figure 2. Phylogenetic tree based on neighbor-joining analysis of the rDNA internal transcribed spacer sequences of the endophytic fungi obtained from Panax ginseng.
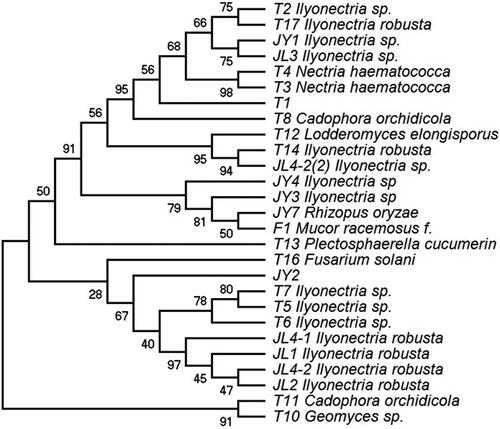
Figure 3. Reaction results of the crude enzyme from ginseng endophytic fungi with pNPG, pNPAp and pNPAf. (A) The reaction results of substrates and crude enzymes extracted from endophytic fungi. (B) Venn diagram of substrates and crude enzyme extracts from endophytic fungi (http://jvenn.toulouse.inra.fr/app/example.html).
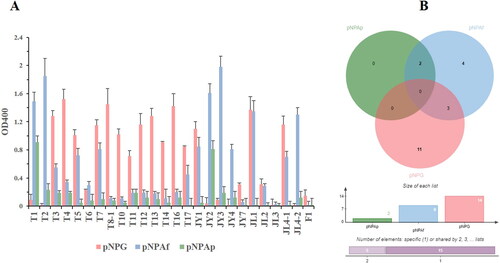
Figure 4. Vector construction of pCold-BglNh. (A) Plasmid digestion with endonuclease. Lane 1: molecular weight standard (cat. # 3428A, Takara); Lane 2: pCold-BglNh vector; Lane 3: pCold-BglNh vector digested by Kpn I restriction endonuclease; Lane 4: pCold-BglNh vector digested by Kpn I and Hind III restriction endonucleases;5, BglNh PCR product (1452 bp). (B) Diagram of pCold-BglNh vector.
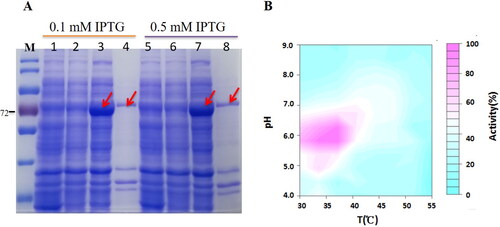
Figure 5. Expression of glycosidase BglNh and determination of optimal reaction conditions. (A) Expression of BglNh. M: Protein Marker (cat. # 26616, Thermo); Lanes 1, 5: Control; Lanes 2,6: before induction; Lanes 3,7: Induced supernatant; Lanes 4,8: Induced precipitate. (B) Contour plot of BglNh using the pNPG as substate to evaluate temperature vs. pH.
Data availability statement
Data and materials related to this work are available from the corresponding author [YL] upon request.