Figures & data
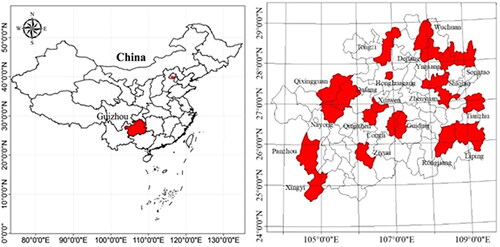
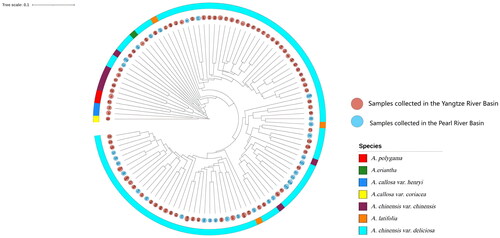
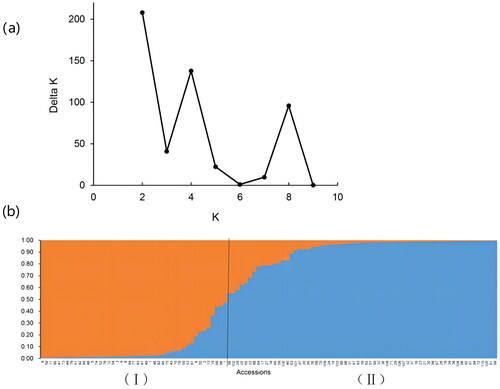
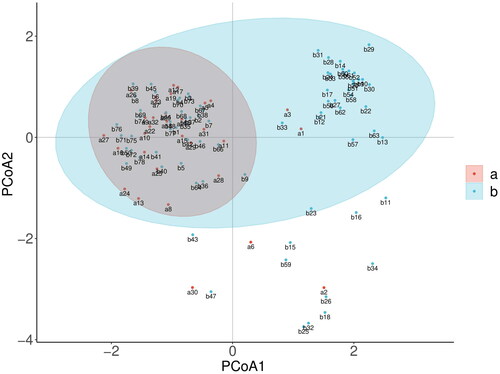
Table 1. Kiwifruit germplasm resources used in this study.
Table 2. The SRAP primers used in the test.
Table 3. SRAP primers used in this study and their polymorphisms.
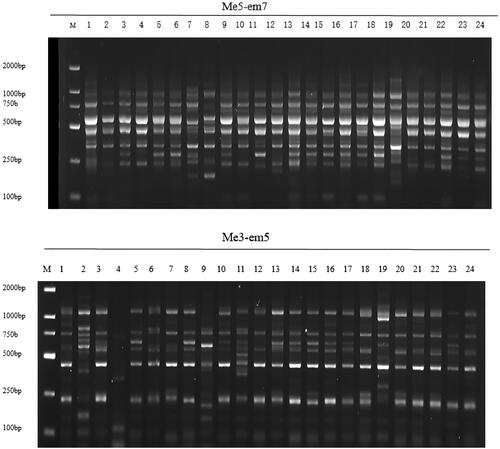
Figure 2. Amplification map generated by several SRAP primers in No. 1–24 kiwifruit materials. M is DL2000 DNA marker (MD114, Tiangen, Beijing, China).
Data availability statement
All data are available from the corresponding author [WL] upon reasonable request.