Figures & data
Figure 1. High-performance liquid chromatogram profile representing several components of γ-oryzanol (A to D) extracted from brown rice.
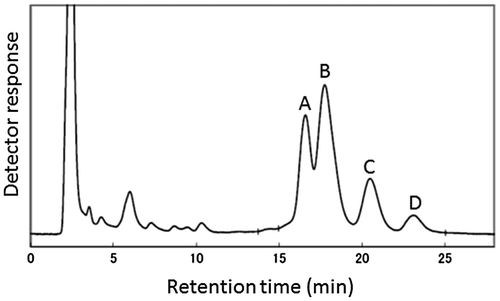
Figure 2. Frequency distributions of total γ-oryzanol content of brown rice among (A) recombinant inbred lines from Asominori/IR24 and (B) backcross inbred lines from Sasanishiki/Habataki//Sasanishiki. Horizontal bar indicates SD.
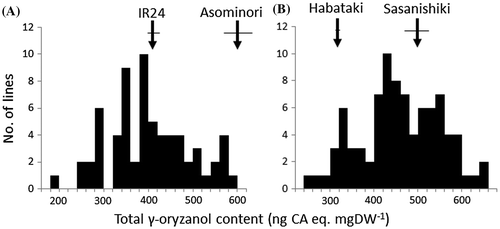
Table 1. QTLs for total γ-oryzanol content in brown rice detected from a set of recombinant inbred lines from Asominori/IR24 and that of backcross inbred lines from Sasanishiki/Habataki//Sasanishiki.
Figure 3. Total γ-oryzanol content of brown rice in chromosome segment substitution lines (CSSLs) with IR24 segments in Asominori background (AIS), and those with Asominori segments in IR24 background (IAS), together with their parents. White, black and hatched bars indicate CSSLs with no significant differences from Asominori, from IR24 and with significant difference from the respective background parents (p < .001), respectively.
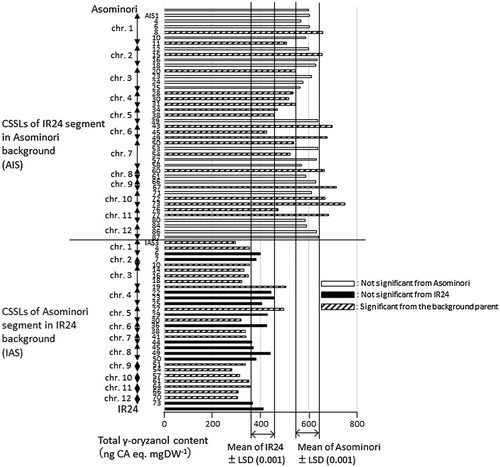
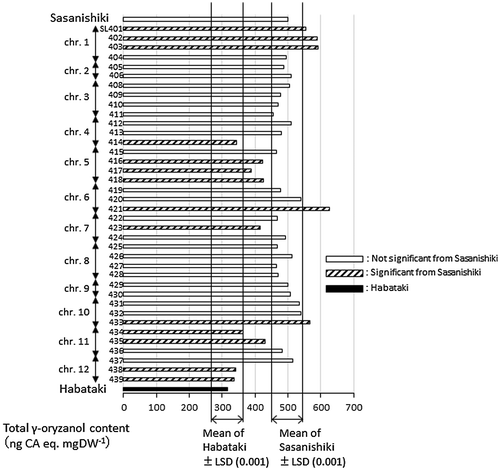
Figure 4. Total γ-oryzanol contents of brown rice in chromosome segment substitution lines (CSSLs) with Habataki segments in Sasanishiki background (SL), together with their parents. White, black and hatched bars indicate CSSLs with no significant differences from Sasanishiki, from Habataki and with significant difference from background parent, Sasanishiki (p < .001), respectively.