Figures & data
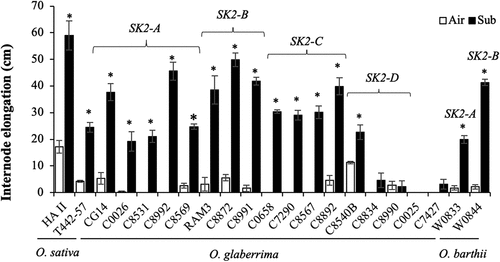
Table 1. Percentage amino acid identity of SK2 in Oryza sativa (Os), Oryza glaberrima (Ogla), Oryza barthii (Obar), and Oryza glumaepatula (Oglu).
Figure 1. Effect of gradual submergence on elongation responses of O. glaberrima and O. barthii accessions and O. sativa deepwater cultivars. Fifty-day-old plants were submerged to a depth of 10 cm for two days, and the water level was then increased at the rate of 5 cm day −1 for 15 days. Air-grown control plants were grown in non-submerged conditions during the same period. (a) Shoot length. The horizontal grey line denotes the final water level (85 cm). (b) Total internode elongation. The SK2 allelic form of each accession of O. glaberrima and O. barthii is shown in the figure. Data represent the mean ± SE of four plants. Asterisks indicate a significant difference (P <0.05, Student’s t-test) between the submerged and control plants.
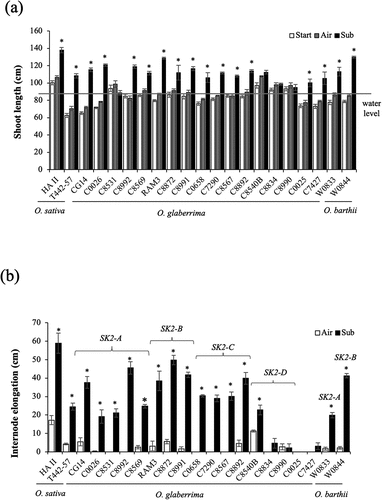
Table 2. Allelic forms of SK,SD1 and ACE1 genes of O. glaberrima and O. barthii accessions and O. sativa deepwater cultivars used in submergence treatment.
Figure 2. Expression analyses of the genes involved in the deepwater response in O. glaberrima. Expressions analyses of SK2 (a), SK1 (b), and SD1 (c) were performed by real-time RT-PCR using gene-specific primers on the internodes of the plants submerged or grown in the air for 12 h. 17s rRNA was used as an internal control. Data represent the means ± SE of three measurements. Different letters above bars indicate significant differences (P <0.05) based on the Bonferroni test. (d) ACE1 expression analysis was performed by RT-PCR using gene-specific primers on the internodes of the plants submerged or grown in the air for 12 h.
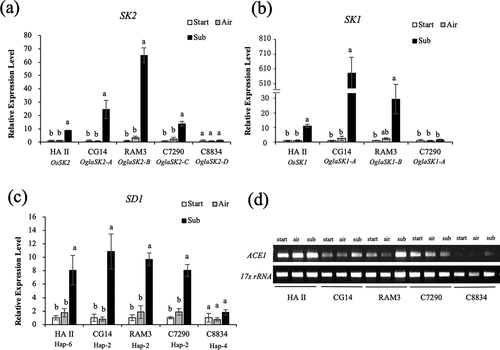
Figure 3. Comparison of internode elongation under submergence in O. glaberrima and O. barthii accessions with different SK1 (a) and SK2 (b) allelic forms. Fifty-day-old plants were submerged to a depth of 10 cm for two days, and then the water level was increased at the rate of 5 cm day −1 for 15 days. Air-grown control plants were grown in non-submerged conditions during the same period. Data represent the means ± SE of 10–23 plants. Different letters above bars indicate significant differences (P < 0.05) based on the Bonferroni test.
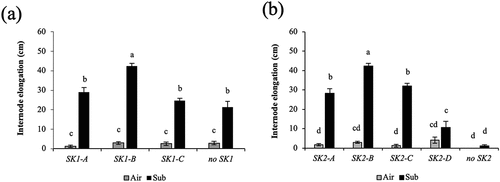
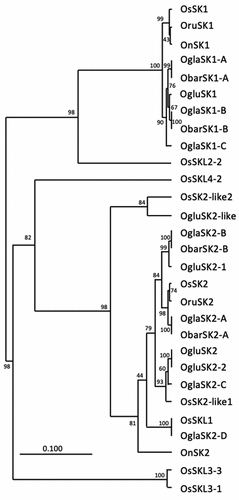
Figure 4. Phylogenetic tree of SK amino acid sequences in Oryza species. Phylogenetic relationships of SK amino acid sequence of Oryza sativa (Os), Oryza rufipogon (Oru), Oryza nivara (On), Oryza glaberrima (Ogla), Oryza barthii (Obar) and Oryza glumaepatula (Oglu) were constructed using the UPGMA method. The bootstrap support values from 1,000 replicants are shown at the node.