Figures & data
Table 1. Clinical parameter and GL gene segments of the included AL amyloidosis patients and sample tissue of extractions.
Figure 1. Location of mutational positions in the patient-specific LC proteins. Sequence alignment of the 10 LCs. Amino acid positions are numbered consecutively. Red: mutation, Green: non-encoded amino acid that arose from junctional diversity, Violet: uncertain position. Underlined amino acids at the C-terminus could not be resolved by cDNA sequencing and represent the amino acids of the respective GL. Above the sequence alignment, the secondary structure elements corresponding to the native structure of a LC (PDB: 6QB6-L) are shown. Arrows indicate β strands and continuous lines indicate ordered conformation. Below the sequence, number of mutations per amino acid position is plotted. Asterisks indicate a mutated position in more than 50% of all LCs. The sequences shown are composed of cDNA sequencing and MS results. Sequences corresponding to FOR005 and FOR006 were already published [Citation28].
![Figure 1. Location of mutational positions in the patient-specific LC proteins. Sequence alignment of the 10 LCs. Amino acid positions are numbered consecutively. Red: mutation, Green: non-encoded amino acid that arose from junctional diversity, Violet: uncertain position. Underlined amino acids at the C-terminus could not be resolved by cDNA sequencing and represent the amino acids of the respective GL. Above the sequence alignment, the secondary structure elements corresponding to the native structure of a LC (PDB: 6QB6-L) are shown. Arrows indicate β strands and continuous lines indicate ordered conformation. Below the sequence, number of mutations per amino acid position is plotted. Asterisks indicate a mutated position in more than 50% of all LCs. The sequences shown are composed of cDNA sequencing and MS results. Sequences corresponding to FOR005 and FOR006 were already published [Citation28].](/cms/asset/c2a659dc-9f1b-420d-81e5-809ceae26774/iamy_a_2095618_f0001_c.jpg)
Figure 2. Overview of the sequence coverage of the fibril proteins of the 10 AL patients found by MS. Disulphide bonded cysteine is highlighted in yellow and pyroglutamate is highlighted in violet. The dashed lines indicate the N- and C-terminal ends.
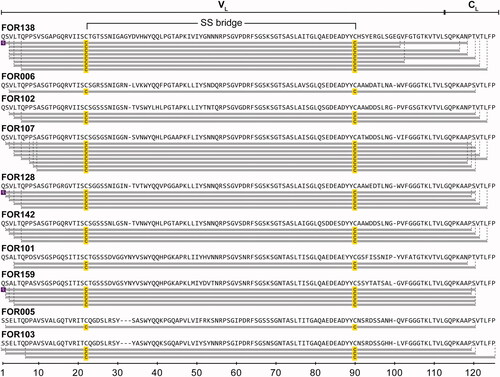
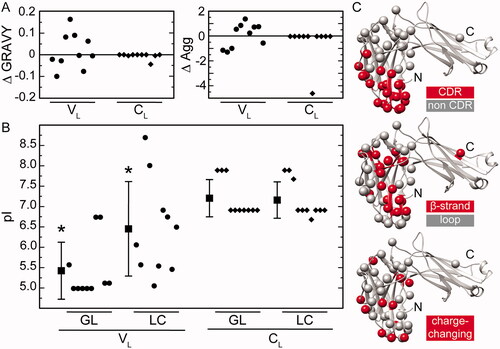
Figure 3. Mutational effects on the biochemical and structural properties of the LC proteins. (A) Analysis of the hydrophobicity (GRAVY) and aggregation propensity (AGG). Shown are the difference values ΔGRAVY and ΔAGG between the LC proteins and their respective GL proteins separately for the VL and CL. (B) Analysis of the pI values. Shown are the individual pI values and the mean pI values of the LC proteins and their respective GL proteins separately for the VL and CL. Asterisks indicate a significant difference. (C) Location of mutations in a natively folded LC (PDB: 6QB6). Colour coding as indicated.
Supplemental Material
Download MS Word (258.9 KB)Data availability statement
The data that support the findings of this study are available from the corresponding author, C. H., upon reasonable request. In addition, the MS data are accessible in the MassIVE database under the dataset identifier MassIVE MSV000089772.
