Figures & data
Table 1. Quantitative RT-PCR primer sequences.
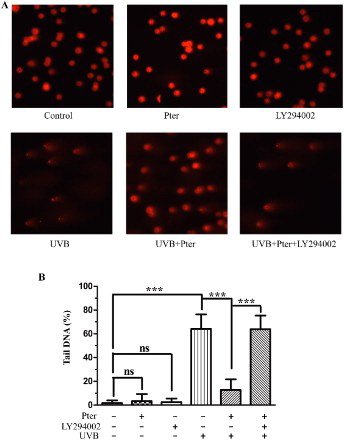
Figure 1. Nrf2 silencing abolished the protective effect of Pter. (A) Viability of HaCaT cells incubated for 24 h in different Pter concentrations. Data are presented as percentage of the untreated control ± SD (n = 6); ns = P > 0.05, ***P < 0.001. (B) Pter-induced Nrf2 nuclear accumulation. HaCaT cell treatment with Pter for 24 h. The indicated proteins were detected by western blotting. GAPDH and lamin A were used to confirm the purity of the cytosolic and nuclear extracts, respectively. (C) Relative protein level of Nrf2. Quantitative densitometric data were expressed as fold change, the vehicle was set to 1. Values are mean ± SD (n = 3); *P < 0.05, ***P < 0.001. (D) Nrf2 was silenced by transfecting with siRNA. HaCaT cells were transfected with a specific siRNA against Nrf2 (siNrf2) or a non-silencing control (NC-siRNA). The knocking down efficiency was demonstrated by western blotting using GAPDH as a loading control. (E) Quantification densitometric data from (D). Values are mean ± SD (n = 3); ns = P > 0.05, ***P < 0.001, compared with non-transfection control. (F) Cell viability in HaCaT cells treated as indicated. Data are presented as percentage of the untreated control ± SD (n = 6); ns = P > 0.05, ***P < 0.001.
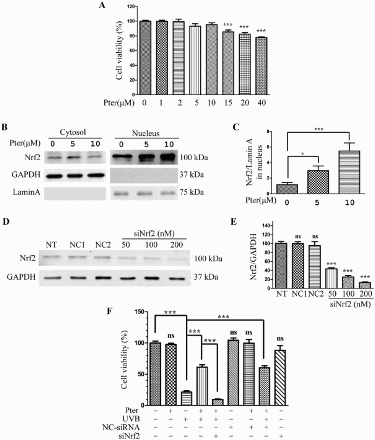
Figure 2. Pter activated Nrf2/ARE via PI3K. After transfection with luciferase reporter plasmid for 24 h, HaCaT cells were pretreated for 1 h with LY294002 prior to Pter treatment. (A) Luciferase activity data are presented as percentage of the untreated control ± SD (n = 6); ns = P > 0.05, ***P < 0.001. (B) The indicated proteins were detected by western blotting, with laminA as the loading control. (C) The levels of phosphorylated Nrf2 were quantified by densitometric scanning. Values are mean ± SD (n = 3); **P < 0.01, ***P < 0.001. (D) Cell viability in HaCaT cells treated as indicated. Data are presented as percentage of the untreated control ± SD (n = 6); ns = P > 0.05, *P < 0.05, ***P < 0.001.
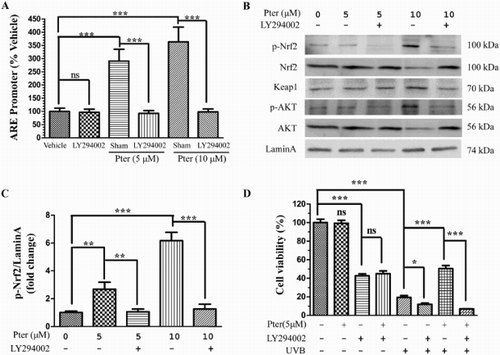
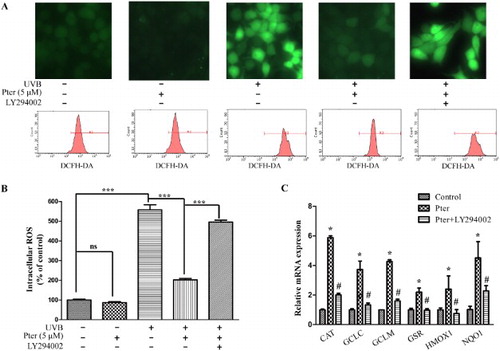
Figure 3. Pter scavenged UVB-induced ROS via PI3K. HaCaT cells were treated with vehicle, Pter, and LY294002 prior to UVB irradiation. (A) Representative intracellular ROS images and recordings. (B) Quantitative fluorescence data from (A) were presented as mean ± SD (n = 3); ***P < 0.001. (C) Quantitative RT-PCR data of indicated Nrf2 target genes. The relative mRNA levels of Nrf2 target genes were represented as fold change; the vehicle was set to 1 with gapdh and actin for normalization. Values are mean ± SD (n = 3), *P < 0.05.