Figures & data
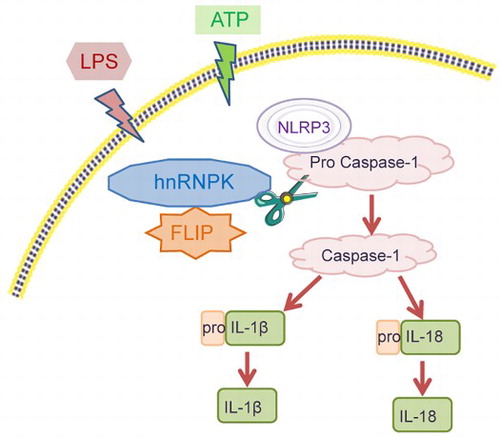
Figure 1. hnRNPK and FLIP were significantly up-regulated in LPS + ATP-activated NLRP3 inflammasome of macrophages. RAW264.7 macrophages were incubated with LPS, followed by ATP incubation (A) Inflammatory factors TNF-α, IL-1β and IL-18 expressions were measured at different time points via ELISA. *p < 0.05, **p < 0.01 vs. Control; #p < 0.05, ##p < 0.01 vs. LPS; (B) The protein expressions of hnRNPK, FLIP, NLRP3, pro Caspase-1, Caspase-1, pro IL-1β, IL-1β, pro IL-18 and IL-18 in different groups were evaluated by western blot. The expression and distribution of (C) hnRNPK, (D) FLIP and (E) NLRP3 were determined in four groups were detected via immunofluorescence.
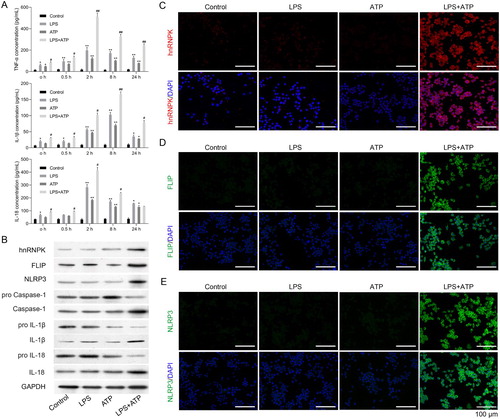
Figure 2. hnRNPK knockdown decreased the expressions of FLIP, NLRP3, Caspase-1, IL-1β and IL-18 in LPS + ATP-activated NLRP3 inflammasome. (A) RAW264.7 macrophages were transfected with three different siRNAs targeting hnRNPK, followed by incubation with LPS and ATP, the mRNA and protein expressions of hnRNPK were detected by real-time PCR and western blot. **p < 0.01 vs. si-NC. RAW264.7 macrophages were transfected with sihnRNPK-2, followed by incubation with LPS and ATP, the mRNA expressions of hnRNPK, FLIP, NLRP3 (B), as well as Caspase-1, IL-1β, IL-18 (C) were detected by real-time PCR. ##p < 0.01 vs. Control, **p < 0.01 vs. LPS + ATP + si-NC.
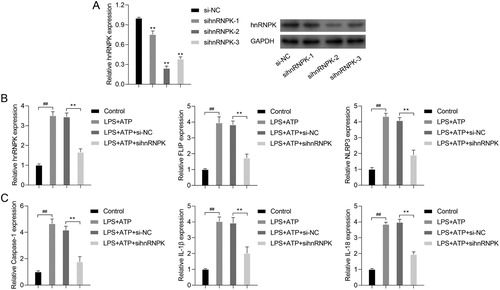
Figure 3. hnRNPK knockdown decreased activated NLRP3 inflammasome related protein levels and bound to FLIP. RAW264.7 macrophages were transfected with si-hnRNPK, followed by incubation with LPS and ATP. (A) The protein expressions of hnRNPK, FLIP, NLRP3, pro Caspase-1, Caspase-1, pro IL-1β, IL-1β, pro IL-18 and IL-18 were evaluated by western blot. The expression and distribution of (B) hnRNPK, (C) FLIP and (D) NLRP3 were determined via immunofluorescence. (E) The binding relationship between hnRNPK and FLIP were detected by co-IP.
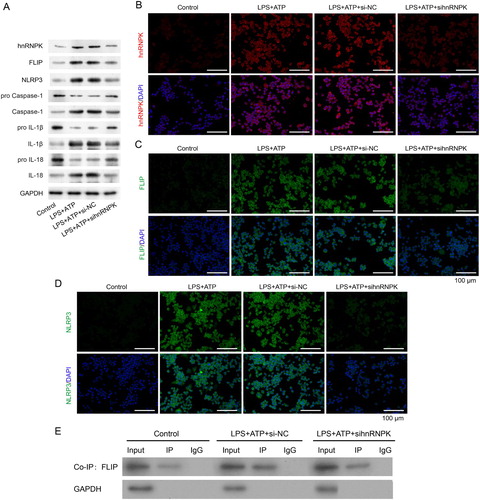
Figure 4. FLIP knockdown decreased the expressions of FLIP, NLRP3, Caspase-1, IL-1β and IL-18 in LPS + ATP-activated NLRP3 inflammasome. (A) RAW264.7 macrophages were transfected with three different siRNA targeting FLIP, followed by incubation with LPS and ATP. the mRNA and protein expressions of FLIP were detected by real-time PCR and western blot. **p < 0.01vs. si-NC. RAW264.7 macrophages were transfected with si-FLIP-3, followed by incubation with LPS and ATP, the mRNA expressions of FLIP, NLRP3 (B), as well as Caspase-1, IL-1β, IL-18 (C) were detected by real-time PCR. ##p < 0.01 vs. Control, **p < 0.01 vs. LPS + ATP + si-NC.
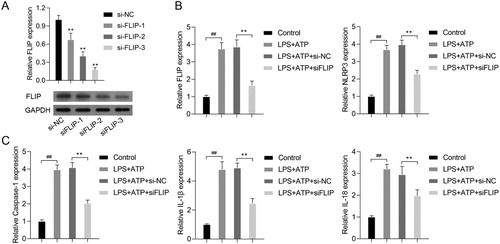
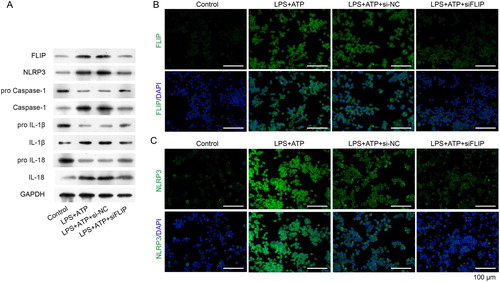
Figure 5. FLIP knockdown decreased activated NLRP3 inflammasome related protein levels. RAW264.7 macrophages were transfected with si-FLIP, followed by incubation with LPS and ATP. (A) The protein expressions of FLIP, NLRP3, pro Caspase-1, Caspase-1, pro IL-1β, IL-1β, pro IL-18 and IL-18 were measured by western blot. (B–C) The expression and distribution of FLIP and NLRP3 were detected by immunofluorescence staining assay.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article.
