Figures & data
Figure 1. Pedigree representing both parents, maternal aunt (identical twin sister) and children. The proband, indicated by the arrow, has complex microphthalmia and a heterozygous SOX2 c.70_89del variant. The mother has a presumed forme fruste coloboma and a dotted shading to denote suspected mosaicism
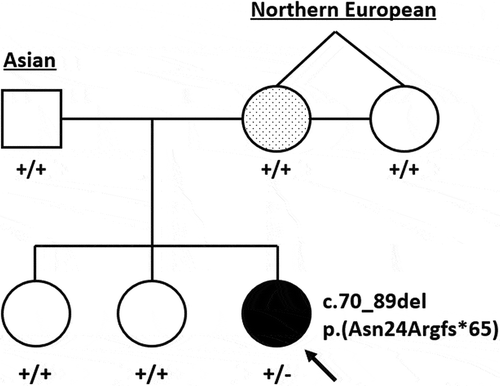
Figure 2. (a) Slit lamp image of the proband’s right eye, showing a vascularized microcornea and a shallow, formed anterior chamber. (b) Proband’s left anterior segment also has microcornea and a mild dyscoria (pupil notch at 3:30 hour). (c) Proband’s posterior segment could only be evaluated OS, showing a uveal coloboma around the optic disc. D) Presumed forme fruste coloboma in the proband’s mother, appearing as iris transillumination patches at 5 o’clock OD and 6 o’clock OS, as well as abnormal pupil dilation OS
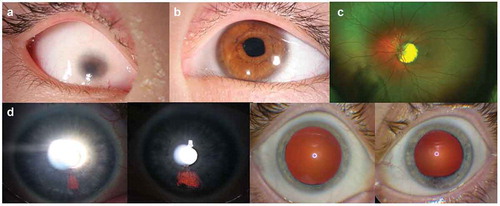
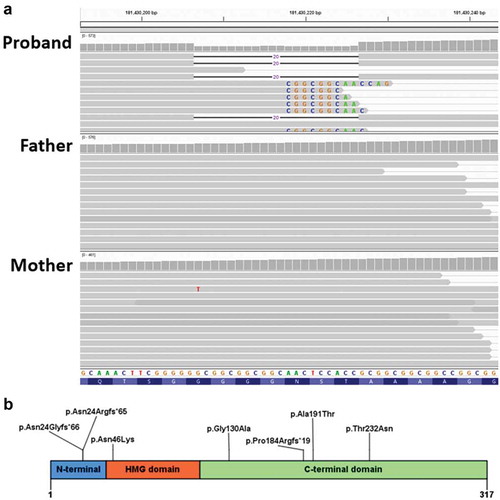
Figure 3. (a) Coverage and alignment tracks in the IGV browser. No read with the 20-bp deletion was found the parents’ data. Sequences of mismatched bases are shown. The reads with mismatched CGGCGGC are expected to contain the 20 nucleotides deletion due to misalignment because of the repeat in the reference sequence. (b) Previously reported predicted SOX2 protein familial variants