Figures & data
Table 1. Chromatographic fractions of petroleum ether extract of A. sativum Linn (Alliaceae).
Table 2. Purification process of the petroleum ether extract of A. sativum with information on yield, specific inhibition activity and purification fold.
Table 3. The EI-Mass, 1H-NMR and antimycobacterial activity by plate assay and MABA of the subfractions F3, F4 and F5.
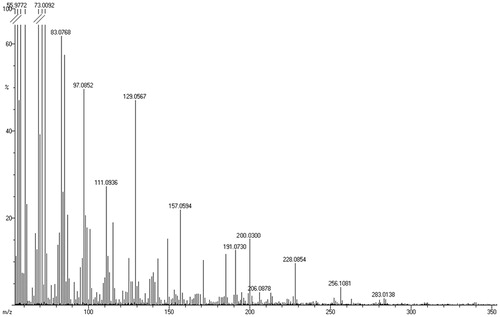
Figure 2. (a) IR spectrum, (b) EI-MS, (c) COSY (expanded spectrum), (d) HSQC (expanded spectrum) in CDCl3 for the bioactive subfraction (F3).
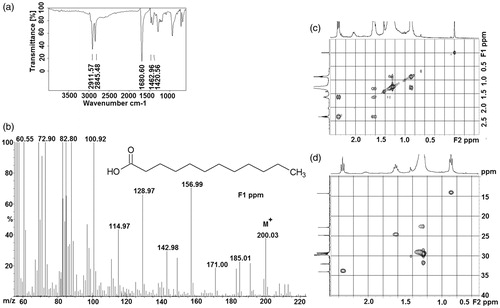
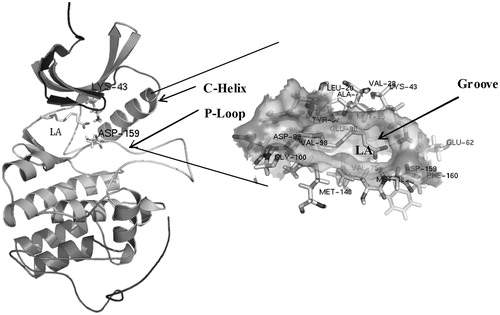
Figure 3. Superposition of the modeled cytosolic domain of PknB on the loop-deficient structure available at the PDB (PDB ID: 1MRU). The modeled loop region, present in between the N-terminal and C-terminal lobe is indicated.
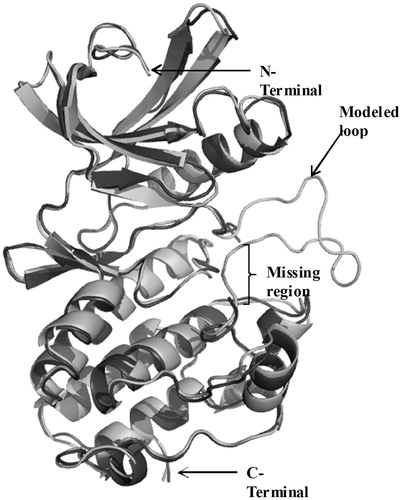
Table 4. The protein IDs, gene name and PDB codes for 12 membrane proteins are shown.
Table 5. Results of lauric acid docking in PknB modeled structure (modeled 1MRU) and PstP (pdb code 2CM1) using Autodock 4.0 software.
Table 6. Binding site residues traced from the ligand with the distance of 4 Å using Swissprot deep viewer.