Figures & data
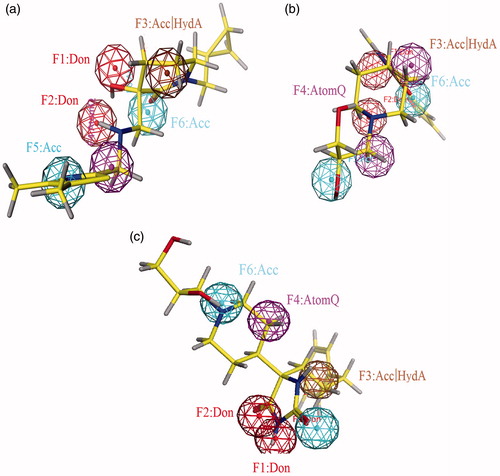
Figure 2. (a) Crystal structure of the PfDXR in complex with inhibitor; (b) Essential chemical features for the generation of pharmacophore; (c) Schematic 2D view of the interactions between fosmidomycin derivative and binding pocket residues; (d) Three dimensional six point pharmacophore model generated by MOE.
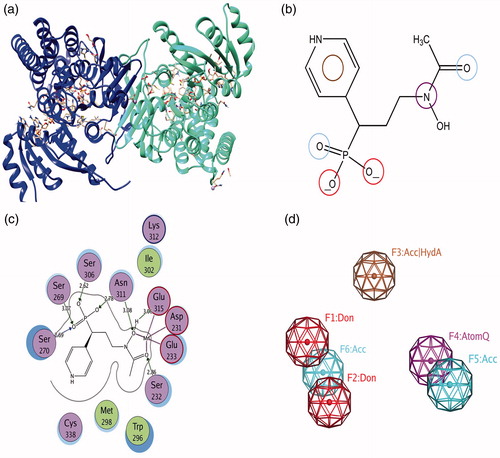
Table 1. 2D structures, ChemBridge database ID, Docking Scores (S), binding energies Kcal/mol, binding affinities Kcal/mol of retrieved hit compounds.
Figure 4. The three dimensional binding mode analysis of most active and least active compounds into the active binding site of PfDXR (a) Hit compound 1; (b) Hit compound 2; (c) Hit compound 13; (d) Hit compound 14.
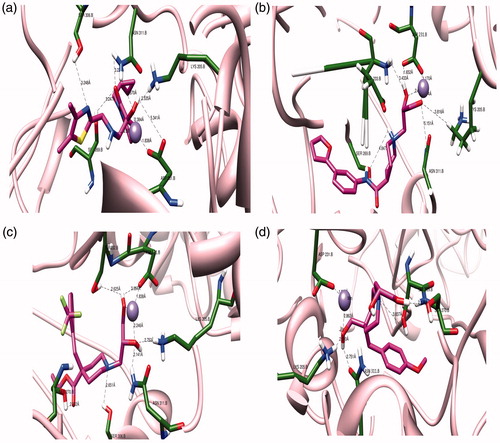
Table 2. Pharmacokinetic and pharmacodynamic descriptors of HITS.
Figure 5. (a) Correlation between predicted Plasma protein binding (Log K) values and logD values: (b) Central nervous system CNS penetration criteria for compounds.
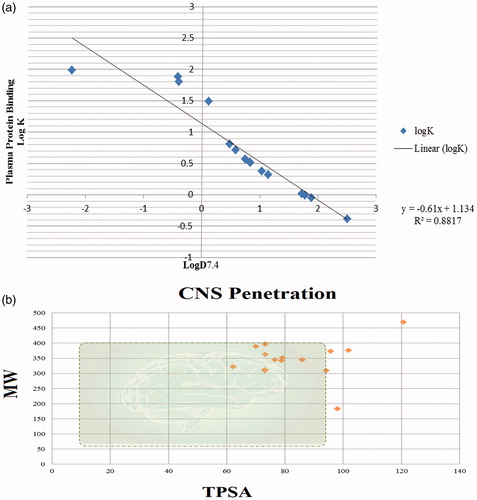
Figure 6. GPCR liabilities of hit compounds. It shows that compounds pertaining pKa> 7 are more substrate at GPCR ligands.
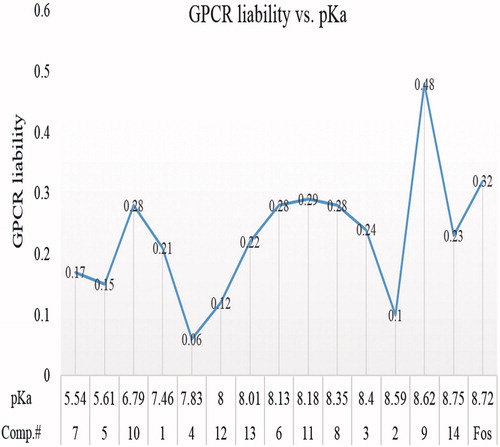
Figure 7. (a) The binding model of compound 4 into the binding site of PfLDH (PDB ID 1LDG): (b–c) The binding model of compound 4 into the binding site of pfDHODH (PDB ID 3O8A): Schematic 2D view of the interactions compound 4 into the binding site of PfDHFR-TS: (d) double mutant (PDB ID 1J3J); (e) quadruple mutant (PDB ID 1J3K) and (f) Wild type inhibitor (PDB ID 1J3I).
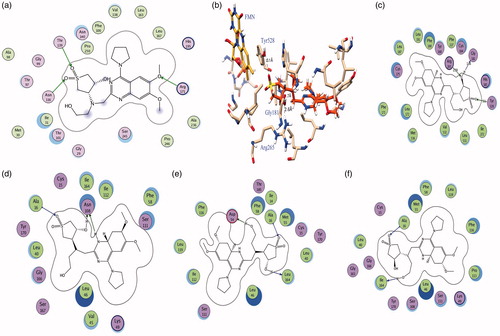
Table 3. Bioactivity predictions and liabilities of designed compounds towards various G-protein coupled receptors GPCRs, ion channels, kinases, nuclear receptors, proteases and various enzymes were estimated using the free online software of molinspiration.
Table 4. Different targets of human malarial parasite P. falciparum and their PDB IDs.
Table 5. MOE docking scores (in Kcal/mol) of retrieved hits showing promiscuity binding with different P. falciparum targets.