Figures & data
Figure 1. The cell viability of tumour cells and NK cells. (A) A549, (B) H1299 and (C) NK cells. Data represent mean ± S.E.M. *p< 0.05; **p< 0.01; ***p< 0.001.
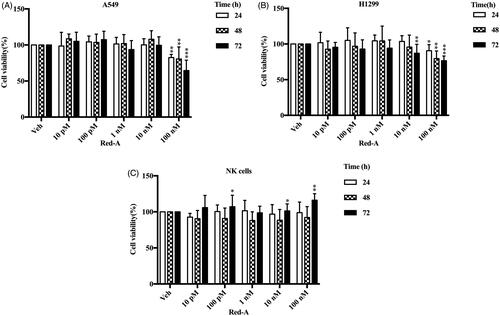
Figure 2. Red-A enhances NK cell-mediated killing of tumour cells. (A, B) The biophotonic cytotoxicity assay and (C, D) impedance assay were performed. E:T represents the ratio of effector cells vs. target cells. Data were pooled from three donors. Data represent mean ± S.E.M. *p< 0.05; **p< 0.01; ***p< 0.001.
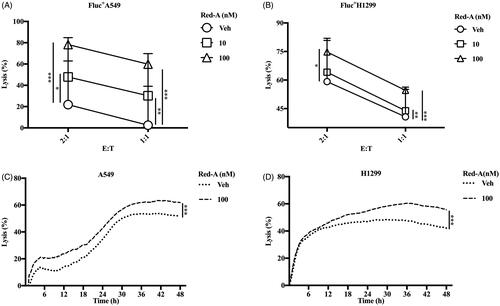
Figure 3. Red-A increases degranulation of NK cells. (A, B) Pre-treated A549 or H1299 cells were co-cultured with NK cells while (C, D) pre-treated NK cells were co-incubated with A549 or H1299 tumour cells. (A, C) Data represent three independent experiments while data (B, D) were pooled from three independent experiments. Data represent mean ± S.E.M. **p< 0.01. NS: non-statistical significance.
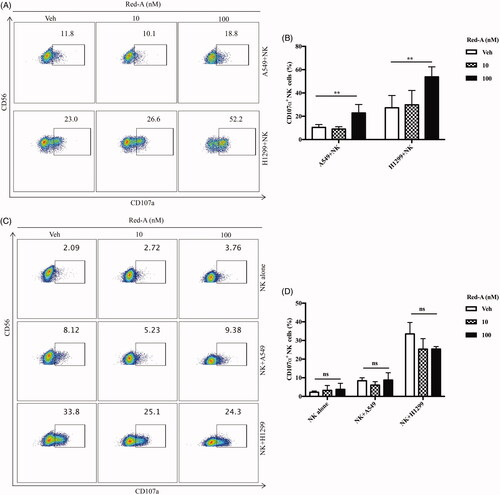
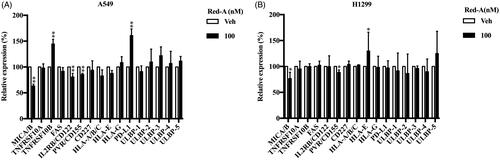
Figure 4. Red-A increases the secretion of IFN-γ. (A, B) NK cells were co-incubated with A549 or H1299 tumour cells in the presence or absence of Red-A, and then IFN-γ was detected by ELISA assay. Data were pooled from three independent experiments. Data represent mean ± S.E.M. *p< 0.05; **p< 0.01.
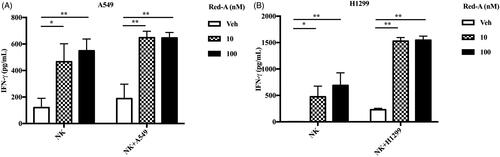
Figure 5. Red-A increases the releasing of granzyme B. (A, B) A549 or H1299 cells were co-cultured with NK cells in different concentrations of Red-A, then granzyme B level in A549 and H1299 tumour cells was assessed. Data A are a representative of three independent experiments while data B were pooled from three independent experiments. Data represent mean ± S.E.M. ***p< 0.001.
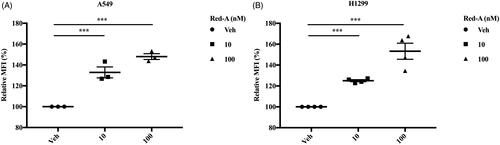
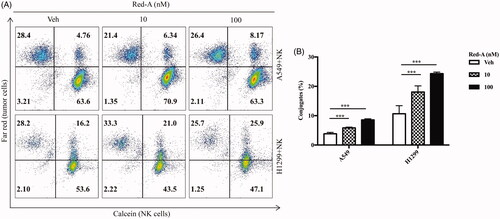
Figure 6. Red-A enhances the conjugation of NK cells and NSCLC cells. (A, B) The population of Far Red/calcein double positive cells was detected by flow cytometry to determine the conjugation of NK cells and tumour cells. Data A are a representative of three independent experiments while data B were pooled from three independent experiments. Data represent mean ± S.E.M. ***p< 0.001.