Figures & data
Figure 1. Chemical structure of rosmarinic acid (RosA). Molecular formula: C18H16O8; molecular weight: 360.32.
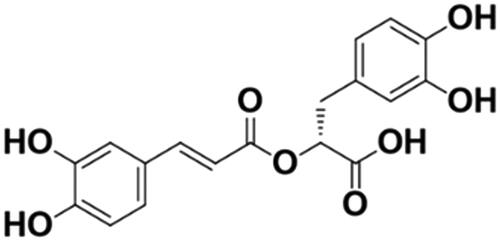
Table 1. Primers for Ogdh and Gapdh showed in the table.
Figure 2. Effects of RosA on myocardial infarct size. (A) Representative photomicrographs of heart sections stained with Evans blue and TTC for different treatment groups are shown. (B) AAR/LV was similar among groups. (C) Treatment with RosA markedly reduced the infarct size caused by myocardial I/R injury. Data are expressed as the mean ± S.D. (n = 6). Significance was determined by ANOVA followed by Tukey’s test. **p < 0.01 vs. Vehicle + I/R. AAR: area at risk; LV: left ventricle; IS: infarct size.
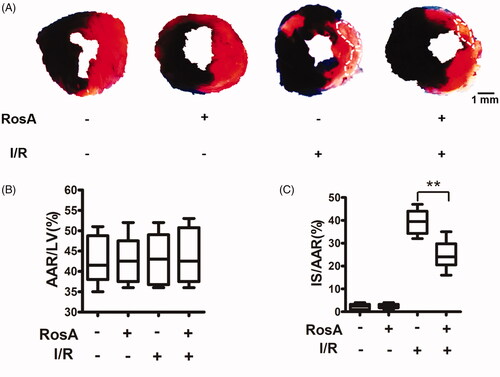
Figure 3. Effects of RosA on cardiac dysfunction. (A) Representative echocardiographs showing cardiac function from the various groups. (B) EF and (C) FS measured by echocardiography. Data are expressed as the mean ± S.D. (n = 4–5). Significance was determined by ANOVA followed by Tukey’s test. **p < 0.01 vs. Vehicle + I/R. EF: ejection fraction; FS: fractional shortening.
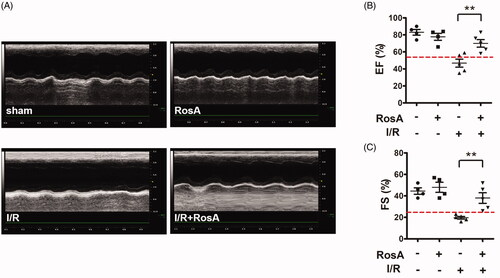
Figure 4. Effect of RosA on levels of CK-MB, cTnI and LDH for each group. (A) RosA reduces CK-MB levels in serum. (B) RosA reduces cTnI levels in serum. (C) RosA reduces LDH levels in culture medium. Data are expressed as the mean ± S.D. (n = 6). Significance was determined by ANOVA followed by Tukey’s test. **p < 0.01 vs. Vehicle + I/R or Vehicle + OGD/R.
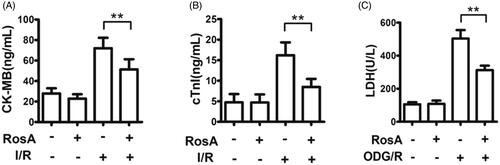
Figure 5. Representative light microscopic images of myocardial histopathological morphology (H&E, ×400).
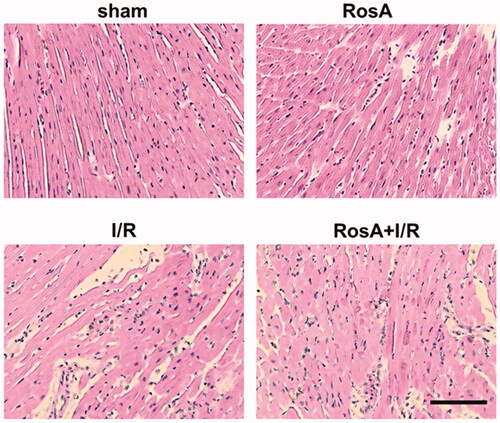
Figure 6. Effects of RosA on ROS production. (A) Representative micrograph of RosA reducing ROS production in myocardial I/R area. Scale bar, 100 μm. (B) Representative micrographs of RosA reducing ROS production after OGD/R injury in cells. (C) Statistical results of RosA reducing ROS production in myocardial I/R area. (D) Statistical results of RosA reducing ROS production after OGD/R injury in cells. Data are expressed as the mean ± S.D. (n = 3). Significance was determined by ANOVA followed by Tukey’s test. **p < 0.01 vs. Vehicle + I/R or Vehicle + OGD/R.
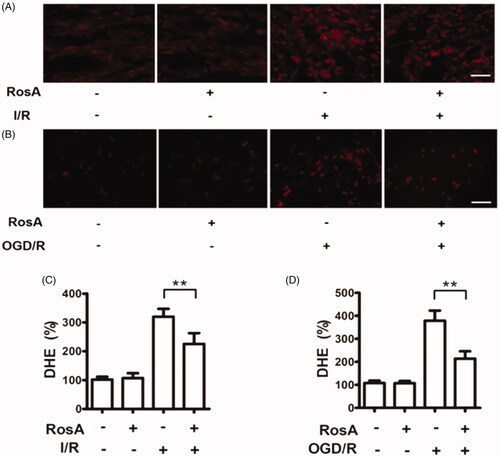
Figure 7. Effects of RosA on oxidative inactivation of ACO. (A) Statistical results of RosA reducing the oxidative inactivation of ACO activity in myocardial I/R area. (B) Statistical results of RosA reducing the oxidative inactivation of ACO activity after OGD/R injury in cells. Data are expressed as the mean ± S.D. (n = 3). Significance was determined by ANOVA followed by Tukey’s test. **p < 0.01 vs. Vehicle + I/R or Vehicle + OGD/R.
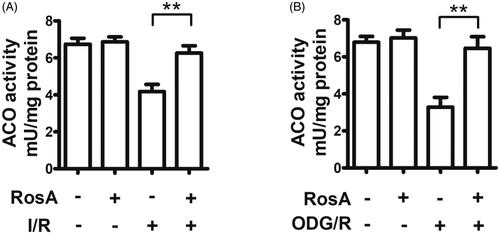
Figure 8. RosA reduced ogdh mRNA and protein levels. (A) RosA reduces ogdh mRNA levels in myocardial I/R area in mice. (B) RosA reduces OGDH protein levels in myocardial I/R area in mice. (C) Statistical results of RosA reducing OGDH protein levels in myocardial I/R area in mice. (D) RosA reduces ogdh mRNA levels after OGD/R injury in cells. (E) RosA reduces OGDH protein levels after OGD/R injury in cells. (F) Statistical results of RosA reducing OGDH protein levels after OGD/R injury in cells. Data are expressed as the mean ± S.D. (n = 3). Significance was determined by ANOVA followed by Tukey’s test. **p < 0.01 vs. Vehicle + I/R or Vehicle + OGD/R.
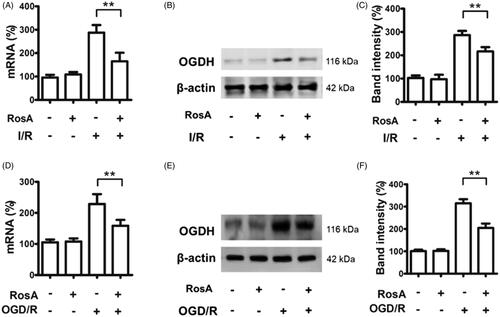
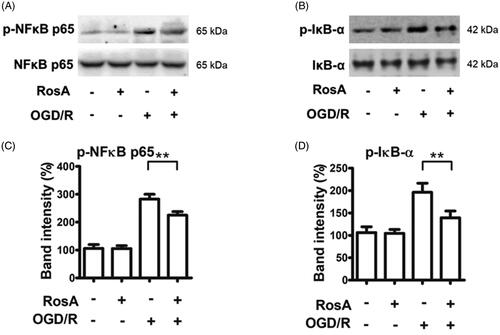
Figure 9. RosA reduced the protein levels of p-NFκB and p-IκB-α. (A) RosA reduced the protein level of p-NFκB. (B) Statistical results of RosA reducing the protein level of p-NFκB. (C) RosA reduced the protein levels of p-IκB-α. (D) Statistical results of RosA reducing the protein level of p-IκB-α. Data are expressed as the mean ± S.D. (n = 3). Significance was determined by ANOVA followed by Tukey’s test. **p < 0.01 vs. Vehicle + OGD/R.