Figures & data
Table 1. Three gRNAs were used to build binary vector pAC0025 targeting CpPDS. Each forward primer was paired with the same critarev reverse primer (atgtacggccagcaacgtcg) to generate PCR amplicons for cloning into the appropriate acceptor vector. Guide RNA target sequences are indicated in all caps within the forward primer sequences. The level 1 acceptor vectors into which PCR products were cloned are indicated, which determine the position of the gRNA cassette in the finished plasmid.
Figure 1. Diagram of the PCR product amplified by primers AC0418 and AC0419. Location of gRNA 1, 2, and 3 target sites, within exons 1 and 2 of CpPDS are indicated.

Figure 2. CRISPR construct T-DNA diagrams. LB indicates left border and RB indicates right border. A) pAC0025 T-DNA includes cassettes for the expression of GFP, selectable marker hptII conferring hygromycin resistance, zCas9i, and three gRNAs targeting CpPDS. B) pAC0026 (the negative control construct) lacks gRNA transcriptional units.
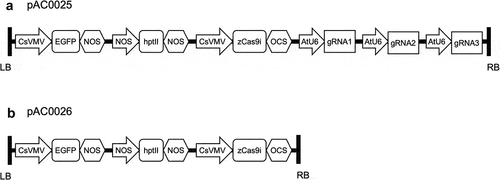
Figure 3. Embryogenic papaya tissue 20 weeks after co-cultivation with Agrobacterium tumefaciens. A) No-selection control showing healthy papaya tissue growing on multiplication media without hygromycin. B) Selection control with untransformed papaya tissue on selective multiplication media supplemented with 50 mg/L hygromycin showing characteristic tissue browning. C) Papaya tissue co-cultivated with Agrobacterium harbouring construct pAC0025 (binary vector with gRNAs targeting CpPDS) with putatively transformed calli actively growing on selective multiplication media circled in red.
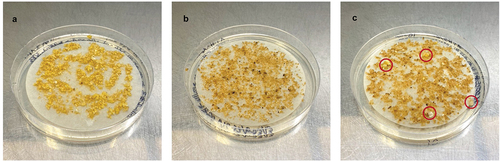
Figure 4. GFP expression in papaya transformed with the plasmid pAC0025. A) Embryogenic papaya culture 34 days post co-cultivation with Agrobacterium. Somatic embryos were primarily at the globular stage. B) Late torpedo (top) and early cotyledon (bottom) stage somatic embryos 131 days post co-cultivation with Agrobacterium.
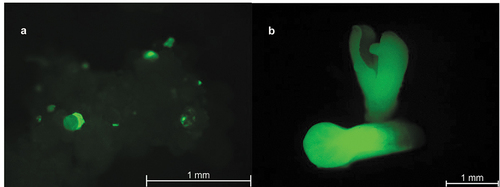
Figure 5. Putative papaya transformants growing on MBN shoot regeneration media with pAC0025 targeting CpPDS (A, B, and C) or negative control vector pAC0026 (D, E, and F). A) pAC0025 transformants immediately after transfer from multiplication media. B) pAC0025 transformants after 4 weeks under lights. C) pAC0025 transformants after 10 weeks under lights. D) pAC0026 transformants immediately after transfer from multiplication media. E) pAC0026 transformants after 4 weeks under lights. F) pAC0026 transformants after 8 weeks under lights.
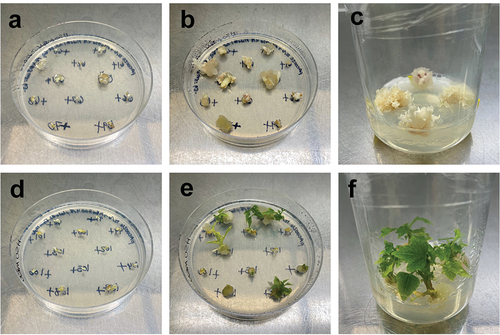
Figure 6. Examples of the range of phenotypes observed in plants transformed with pAC0025, the construct targeting CpPDS. A) Wild-type phenotype (dark green), B) Pale green phenotype, and C) Albino phenotype.

Figure 7. PCR amplification of genomic DNA from putatively edited plants and controls. The first and last lanes in each row were loaded with GeneRuler 1kb+ DNA ladder. Lane labels reflect the source of genomic DNA utilised in each PCR reaction. ‘pAC0025-transformed lines’ indicates PCR reactions utilising gDNA from papaya plants transformed with pAC0025, the CRISPR/Cas9 plasmid targeting CpPDS. Lanes labelled ‘pAC0026 transformant’ indicate PCR with gDNA from a plant transformed with the no-gRNA negative control plasmid pAC0026. Lanes labelled ‘Plasmid pAC0025 DNA’ and ‘Plasmid pAC0026 DNA’ correspond to PCR reactions utilising purified plasmid DNA as template. In ‘Wild-type’ lanes gDNA from an untransformed papaya plant was used as PCR template, and ‘water’ lanes served as a negative control with no DNA template added to the PCR reactions. Partial CpPDS sequence (top row) wild-type amplicon size 909 bp and partial GFP CDS from T-DNA insertion (bottom row) expected amplicon size is 287 bp.
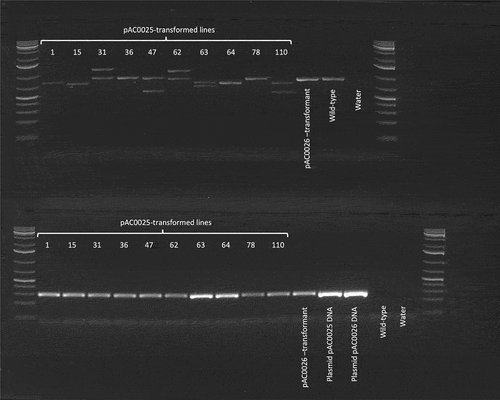
Table 2. CpPDS genotypes of plant lines transformed with pAC0025. “-“ indicates a deletion, ‘+’ an insertion, and ‘inversion’ denotes that the sequence between cut sites has been excised, reversed, and re-ligated. ‘WT’ indicates wild-type sequence, or the absence of mutations.
Table 3. Sequence alignment of CpPDS genotypes at all three gRNA target sites. ‘WT’ indicates wild-type sequence. ‘Al.’ represents allele, described here as ‘A’ or ‘B’. For plant lines found to be homozygous for CpPDS, alleles are identical and represented as ‘A/B’. PAM sites are denoted by underlined characters, italicised sequences represent gRNA targets, “-“ indicates a deletion, ‘+’ an insertion, and grey shading designates an inversion.
