Figures & data

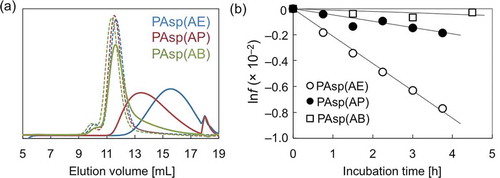
Figure 1. (a) SEC chart showing PAsp(AE), PAsp(AP), and PAsp(AB) before (dotted line) and after incubation for 24 h (solid line) under the physiological condition (pH 7.4, 37 °C); (b) lnf of PAsp(AE) (○), PAsp(AP) (●), and PAsp(AB) (□) plotted against incubation time.
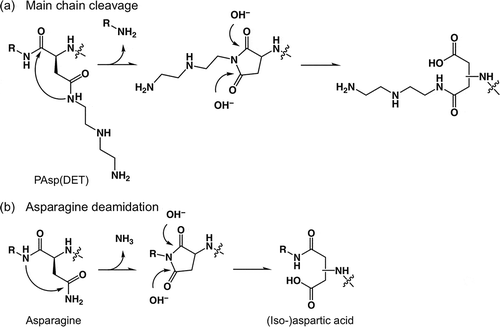
Scheme 2. Synthesis of PAsp(AE), PAsp(AP), and PAsp(AB) by aminolysis of PBLA with corresponding diamino compounds.
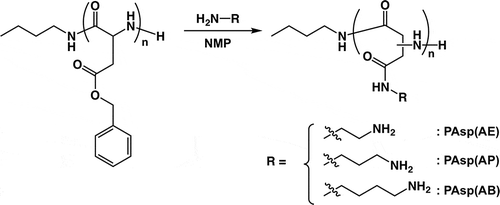
Table 1. Cleavage rate constant (k) [h–1] and T [h] of PAsp(R)s.
Figure 2. (a) Expected conformational changes in the side chain of PAsp(AE); and (b) expected ring-like conformations in the side chain of PAsp(R) series and the stabilization tendency are shown.
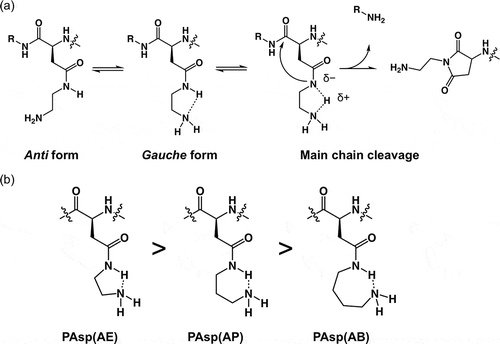
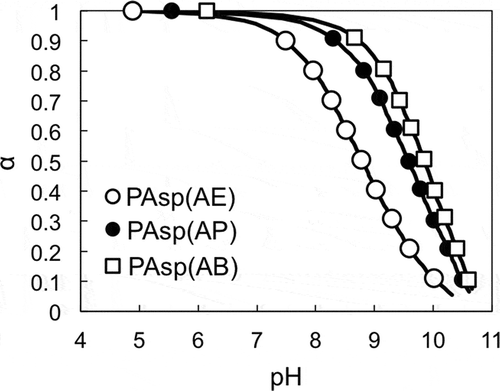
Figure 4. (a) lnf of PAsp(AE) at different pH values; (b) changes in cleavage rate constant (k) as a function of the degree of deprotonation of the primary amines in the side chains of PAsp(AE) (○), PAsp(AP) (●), and PAsp(AB) (□).
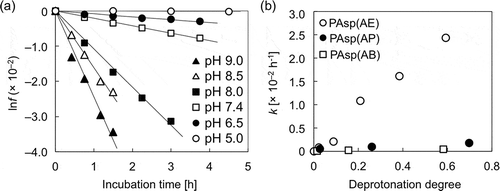
Figure 5. Viability of Huh-7 cells after 48 h incubation with (a) PAsp(AE), (b) PAsp(AP), and (c) PAsp(AB) at varying concentrations. Each PAsp(R) was pre-incubated for 0 or 48 h at 37 °C and pH 7.4. Results are expressed as mean ± SD (n = 5). The data were statistically analyzed by Student’s t-test in Microsoft Excel (*p < 0.0005).
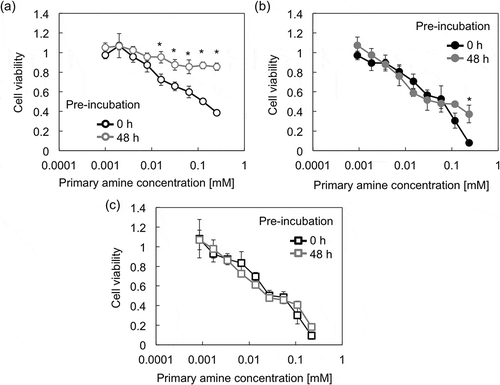
Figure 6. (a) Size distribution histograms of mRNA-loaded PICs prepared from PAsp(AE) (○), PAsp(AP) (●), and PAsp(AB) (□); and (b) changes in SLI of mRNA-loaded PICs after 0 h (white bar), 24 h (gray bar), and 48 h (black bar) incubation at 37 °C and pH 7.4. Results are expressed as mean ± SD (n = 3).
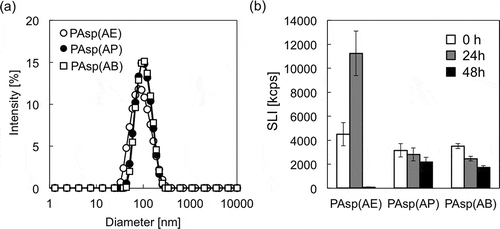
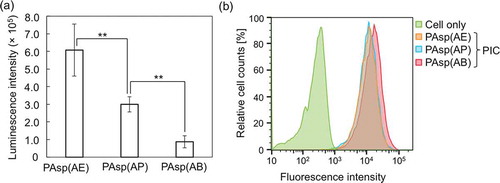
Figure 7. (a) GLuc mRNA expression level in cultured Huh-7 cells treated with GLuc mRNA-loaded PICs. Results are expressed as mean ± SD (n = 5). The data were statistically analyzed by an analysis of variance, followed by Tukey’s post hoc test (**p < 0.01). (b) Flow cytometry peaks showing cellular uptake of Cy5-mRNA-loaded PICs in cultured Huh-7 cells. In both experiments, 250 ng of GLuc mRNA was transfected to Huh-7 cells for 24 h.