Figures & data
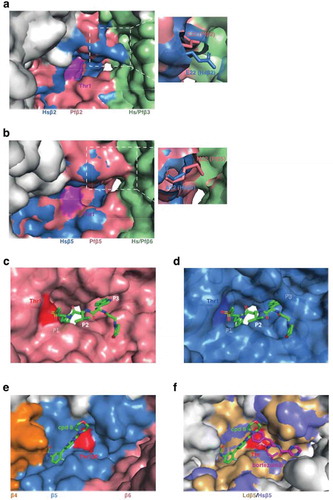
Figure 1. Molecular architecture of the 20S proteasome. (a) Side view of the P. falciparum 20S proteasome (PDB: 6 muw) showing two β-rings sandwiched between two α-rings. (b) The catalytic subunits are highlighted. (c) Cross-section of the P. falciparum 20S core showing the catalytic chamber with β1 and β2 (top β-ring) and β5 (bottom β-ring) active sites indicated. In the absence of activator, the α subunit gate is closed. (d) Residues (P1, P2, P3 and P1ʹ, P2ʹ, P3ʹ) on either side of target amide bond in the protein substrate interact with active sites in the proteasome core via substrate binding pockets (termed S1, S2, S3 and S1ʹ, S2ʹ, S3ʹ). (e) The gate to the 20S core is opened by the binding of regulators, including the ubiquitin-dependent, 19S regulatory particle, and the ubiquitin-independent, PA28 activator. (f) Modeled structures (space-filling representation of resolved residues) for the unbound and PA28-bound α-ring of the P. falciparum 20S proteasome (PDB: 6 muw) showing the blocked (left) and opened (right) central pore.
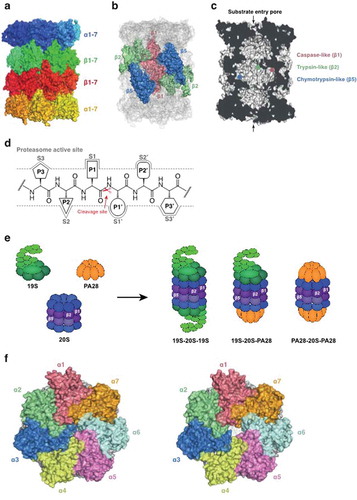
Figure 2. Some examples of proteasome inhibitors. The epoxyketone, Carmaphycin B analogue 18 (a) and the vinyl sulfone, WLL-vs (b) are irreversible covalent inhibitors. The peptide boronate, MPI-1 (c) is a covalent reversible inhibitor. TDI4258, GNF6701 and GSK3494245 (Compound 8) are non-covalent inhibitors.
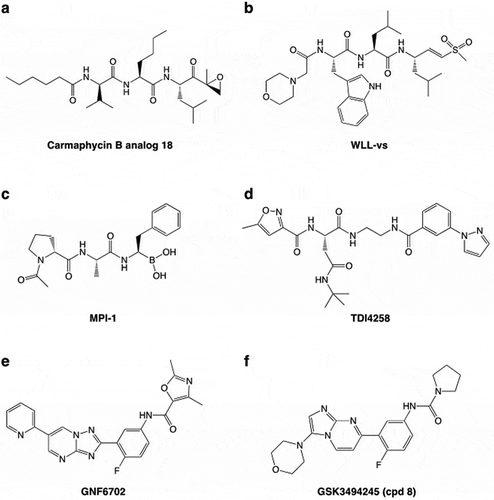
Figure 3. Structural analysis of the proteasome reveals the basis of inhibition. (a and b) Overlays of the Pf20S (PDB 6muw) and human 20S (PDB 5le5) Pf20S β2 (a) and β5 (b) active sites. The active site threonine (Thr1) is shown in purple. The P. falciparum 20S β2 active site is open and can accommodate bulkier substrates. The β5 P3 pocket is narrower due to Met22 replacing the equivalent Ala22 in human β5. (c) CryoEM structure of P. falciparum 20S β2 with bound WLW-vs (PDB 5fmg). The active site threonine is shown in red. (d) The human 20S β2 (PDB 5le5) active site with docked WLW-vs illustrating the poor fit. (e) CryoEM structure of the L. donovani 20S β5 active site (PDB 6qm7). Compound 8 (cpd8) binds to the space between Ld20S β4 and β5 subunits and close to the active site threonine (red). (f) Overlay of Ld20S/cpd8 (PDB 6qm7; beige) with human 20S structure/bortezomib (PDB 5lf3; purple). Relative binding positions of cpd8 and bortezomib are depicted.