Figures & data
Figure 1 Structure of L-aspartol adenylate (Asp-ol-AMP)(1), a stable analog of the natural intermediate L-aspartyl adenylate (2) of the aminoacylation reaction catalyzed by AspRS.
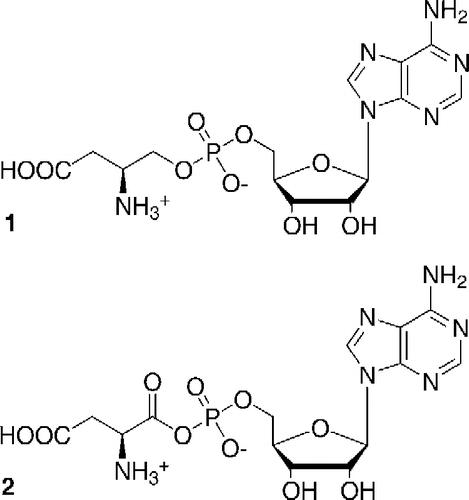
Figure 2 Inhibition by Asp-ol-AMP of pure tRNAAsp and tRNAAsn aspartylation catalyzed by the ND-AspRS of P. aeruginosa. ▾, P. aeruginosa tRNAAsp aspartylation (Ki = 41 μM; r2 = 0.9943); ▪, P. aeruginosa tRNAAsn aspartylation (Ki = 215 μM; r2 = 0.9625). The straight and dashed lines represent the competitive inhibition curves plotted using the above mentioned inhibition constants, for tRNAAsn and tRNAAsp aspartylation, respectively.
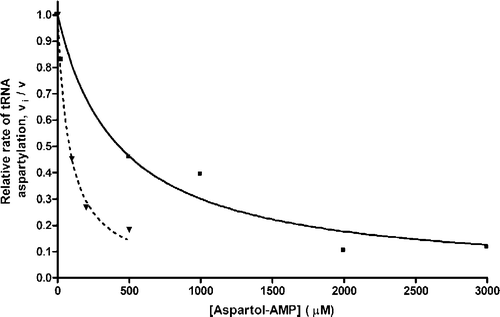
Figure 3 Secondary structures of (a) tRNAAsp and (b) tRNAAsn from P. aeruginosa PAO1. The sequences are derived from the genes, and thus do not show the nucleotide modifications. Two species of tRNAAsp exist in P. aeruginosa, and are only different at base pair number two (C:G or G:C), as shown in (a). The white letters in black circles represent the identity elements of tRNAAsp 16. The differences between these two tRNAs are boxed. Some nucleotides in the anticodon loop are identified by their position number in the standard tRNA structure.
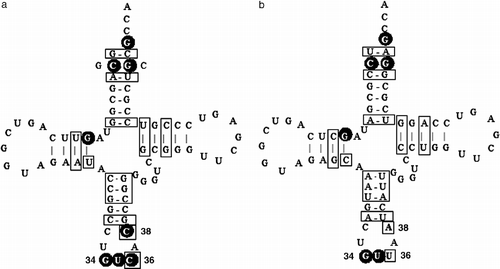
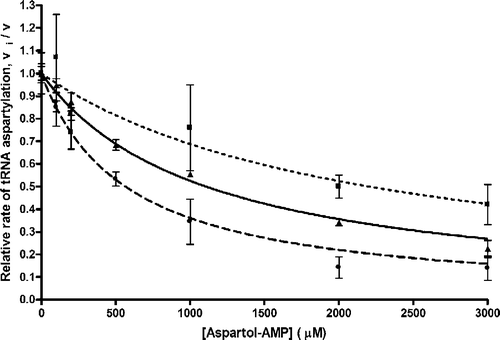
Figure 4 Inhibition by Asp-ol-AMP of yeast – wild-type, and C36U and C38A variants, tRNAAsp transcript aspartylation catalyzed by the ND-AspRS of P. aeruginosa. •, C38A tRNAAsp transcript aspartylation (Ki = 280 μM); ▴, wild-type tRNAAsp transcript aspartylation (Ki = 550 μM); ▪, C36U tRNAAsp transcript aspartylation (Ki = 1100 μM). Lines represent the competitive inhibition curves plotted using the above mentioned inhibition constants. The mean ± spread is shown from duplicate (C36U) and standard error from triplicate (wild-type and C38A) experiments.