Figures & data
Figure 1 Chemical structures of the investigated compounds: caffeic acid (1), rosmarinic acid (2), lithospermic acid B (3), 12-hydroxyjasmonic acid 12-O-β-glucoside (4), and p-menth-3-ene-1,2-diol 1-O-β-glucopyranoside (5).
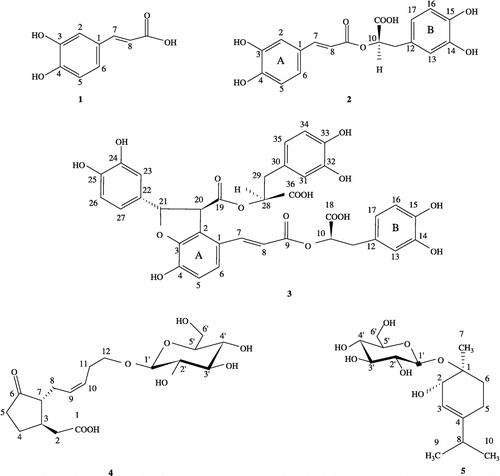
Table I. Inhibition of soybean lipoxygenase in vitro IC50 (mM), calculated binding energy for the highest ranked docking solution and calculated Log P value of the docked conformers.
Figure 3 Conformational comparison between X-ray structure of 13-HPOD (colored by atom type) (PDB IK3) and predicted (colored yellow) by GLUE. Iron atom is illustrated as a sphere (colored magenta). Colour plate can be viewed in the online version.
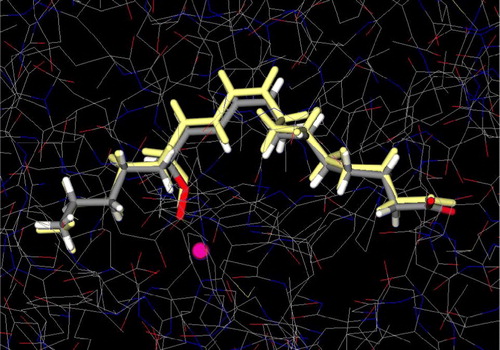
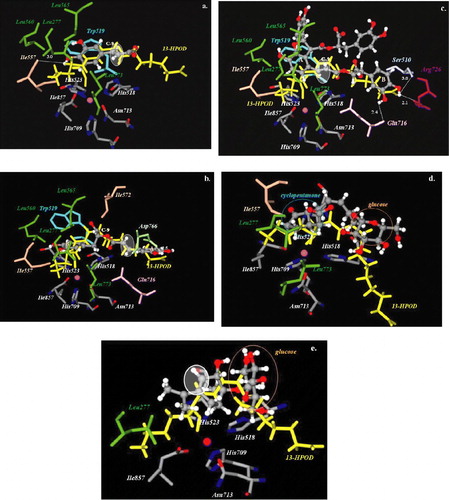
Figure 4 Docked orientations of a. caffeic acid (1) b. rosmarinic acid (2) c. lithospermic acid B (3) d. 12-hydroxyjasmonic acid 12-O-β-glucopyranoside (4) and e. p-menth-3-ene-1,2-diol 1-O-β-glucopyranoside (5) (illustrated as a ball-and-stick model colored by atom type) with additional depiction of selected amino acid residues of soybean lipoxygenase (PDB IK3) as well as 13-HPOD (drawn as sticks). Hydrogen bonds and polar interactions are shown as dotted lines. White colored circles highlight double bond position of the compounds. Iron atom is illustrated as a sphere (colored magenta). Coloured plate can be viewed in the online version.
![Figure 2 Chemical structure of 13-HPOD [9Z,11E-13(S)-hydroxyperoxy-9,11-octadecadienoic acid].](/cms/asset/b2ebb704-0330-475f-b83a-db63aeb4a2a3/ienz_a_199002_f0002_b.gif)