Figures & data
Table I. Structure and CCR5 binding affinities of sulfonyl derivatives of piperidine containing compounds.
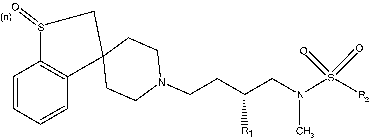
Table II. Structure and CCR5 binding affinities of non-spiro piperidine derivatives.
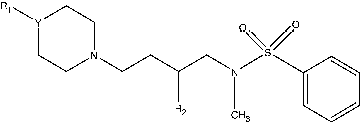
Table III. Structure and CCR5 binding affinities of spiro piperidine derivatives.
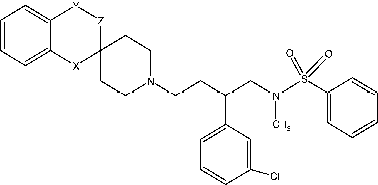
Table IV. Structure and CCR5 binding affinities of piperidine derivatives.
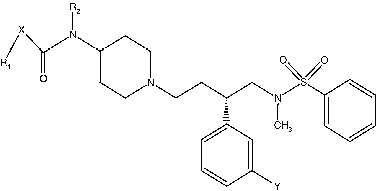
Table V. Categorical list of descriptors used in the development of models.
Table VI. Serial numbers of compounds under different clusters.
Table VII. Comparative study of statistical parameters of classical QSAR models using different descriptors.
Table VIII. Comparative study of statistical parameters of GFA-MLR models using different descriptors.
Table IX. Comparative study of best two network using different hidden layers.
Table X. Comparison of external predictability characteristics of different models obtained from the training set using classical QSAR.
Table XI. Comparison of external predictability characteristics of different GFA-MLR models.
Table XII. Comparison of external predictability characteristics of different ANN models.
Table XIII. Calculated values of k and k/ for different models as defined by Golbraikh and Tropsha [Citation49].
Table XIV. Comparison of best ![]() between observed and predicted values of the test set compounds using different techniques.
between observed and predicted values of the test set compounds using different techniques.
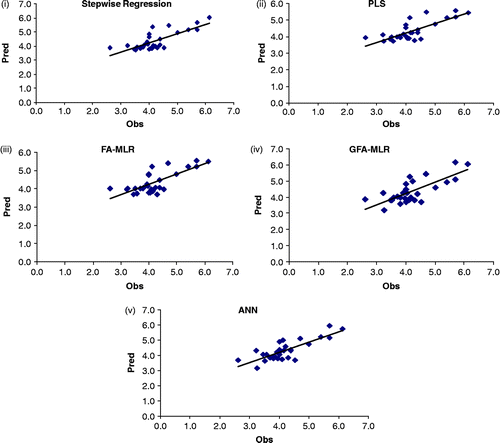
Figure 2. Scatter plots of observed versus predicted values of the test set compounds obtained from the best models (based on rm2 value for the test set compounds) using (i) stepwise regression (Equation 7), (ii) PLS (Equation 8), (iii) FA-MLR (Equation 9), (iv) GFA-MLR (Equation 11) and (v) ANN (6th model).