Figures & data
Figure 1. Studied HIV-PRs and their mutations. (a) Amino acid sequence alignment of the four HIV-PR. The mutations, either polymorphic or arisen from treatment, are marked [Citation9]: primary in black and secondary in grey. (b) Crystallographic structures of the proteases Fwt (PDB code 3P3C), Bmut (PDB code 2P3A) and Fmut (PDB code 2P3D) [Citation29]. Primary mutations are represented by black spheres, secondary by grey spheres, and other differences in white spheres.
![Figure 1. Studied HIV-PRs and their mutations. (a) Amino acid sequence alignment of the four HIV-PR. The mutations, either polymorphic or arisen from treatment, are marked [Citation9]: primary in black and secondary in grey. (b) Crystallographic structures of the proteases Fwt (PDB code 3P3C), Bmut (PDB code 2P3A) and Fmut (PDB code 2P3D) [Citation29]. Primary mutations are represented by black spheres, secondary by grey spheres, and other differences in white spheres.](/cms/asset/b08bee60-110e-4bca-9b14-7c01a6c56b7e/ienz_a_332341_f0001_b.gif)
Figure 2. Chemical structures of the six FDA-approved HIV PR inhibitors used in present study. Schematic representations of APV, amprenavir; IDV, indinavir; LPV, lopinavir; NFV, nelfinavir; RTV, ritonavir; SQV, saquinavir; PPT, acetyl-pepstatin and TL-3 are given.
Figure 3. Inhibition curves of (a) Bwt, (b) Bmut, (c) Fwt and (d) Fmut HIV PRs by amprenavir. V/V0 values have been obtained as described in Materials and Methods. Solid curve represents the fit of Equation (1) to the experimental data. The differences in the concentration ranges of amprenavir used for inhibition of subtype B and subtype F HIV PRs and their drug-resistant mutants should be noted.
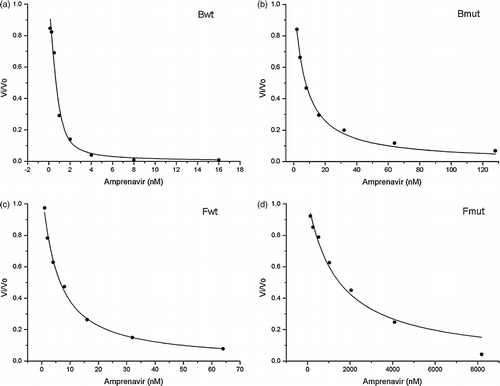
Table I. Catalytic efficiency for all four studied HIV-1 proteases.
Table II. Inhibition constants (Ki) for studied HIV-1 proteases.
Table III. Vitality values for subtype B and F HIV proteases.
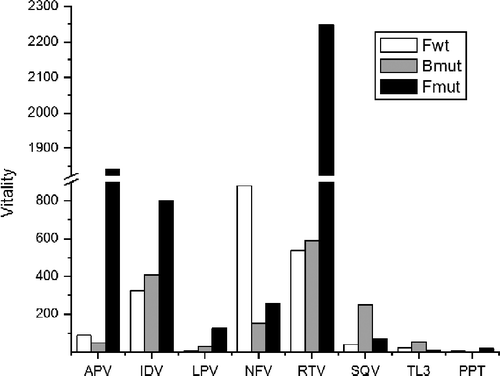
Figure 4. HIV PRs vitality bar plot for all eight protease inhibitors studied for Fwt, Bmut and Fmut proteases (Bwt as reference). The highest vitality values are observed for Fmut in response to amprenavir, ritonavir and indinavir; and for Fwt in the presence of nelfinavir and ritonavir.