Figures & data
Figure 1. Comparison of the structure of TLXI (A, based on [23]) with that of thaumatin (B, based on [26]). The different domains are indicated by I, II and III. Disulfide bridges are shown in light grey. The large ß-sheets shown in light and dark grey form a ß-sandwich. A third smaller ß-sheet is shown on the right hand side of the molecules. The α-helices of thaumatin are shown as right-handed coils. The structures are visualized with Pymol 0.99rev8 [31].
![Figure 1. Comparison of the structure of TLXI (A, based on [23]) with that of thaumatin (B, based on [26]). The different domains are indicated by I, II and III. Disulfide bridges are shown in light grey. The large ß-sheets shown in light and dark grey form a ß-sandwich. A third smaller ß-sheet is shown on the right hand side of the molecules. The α-helices of thaumatin are shown as right-handed coils. The structures are visualized with Pymol 0.99rev8 [31].](/cms/asset/d191b2c4-94a0-461a-95e4-a9594d908439/ienz_a_332350_f0001_b.gif)
Figure 2. Sequence alignment (ClustalW) of short TLPs from barley (TLP 1, 2, 3, 4) [Citation27], wheat (PWIR2) [Citation32] and rice (pPIR2) [Citation33], thaumatin [Citation34] and TLXI. Domain III is indicated with a grey arrow, while domain II is indicated with a black arrow. The rest of the protein constitutes domain I. The conserved cysteine residues are indicated in bold and the symbols above indicate which residues form disulfide bonds.
![Figure 2. Sequence alignment (ClustalW) of short TLPs from barley (TLP 1, 2, 3, 4) [Citation27], wheat (PWIR2) [Citation32] and rice (pPIR2) [Citation33], thaumatin [Citation34] and TLXI. Domain III is indicated with a grey arrow, while domain II is indicated with a black arrow. The rest of the protein constitutes domain I. The conserved cysteine residues are indicated in bold and the symbols above indicate which residues form disulfide bonds.](/cms/asset/49161ec7-8b96-4eaa-ad1d-1f49d08f703d/ienz_a_332350_f0002_b.gif)
Figure 3. Circular dichroism spectrum of TLXI. The spectrum is normalized to the protein concentration and expressed as mean residue ellipticity [θ].
![Figure 3. Circular dichroism spectrum of TLXI. The spectrum is normalized to the protein concentration and expressed as mean residue ellipticity [θ].](/cms/asset/e98ab83e-1d32-4806-a811-a9be2767aebf/ienz_a_332350_f0003_b.gif)
Table I. Analysis of the number of free cysteine residues in one TLXI molecule, determined with Ellmann's reagent. The number found in a TLXI sample treated with sodium borohydride was compared with the number in an untreated TLXI sample.
Figure 4. Residual inhibition activity of TLXI against XTL1 after 2 h incubation at different pH conditions and at room temperature (S.D. ≤ 5.7; n = 3) (A). Residual inhibition activity of TLXI against XTL1 after 40 min incubation at different temperatures and pH 5.0 (S.D. ≤ 3.9; n = 3) (B) and after different times at pH 5.0 and 100°C (S.D. ≤ 9.7; n = 3) (C).
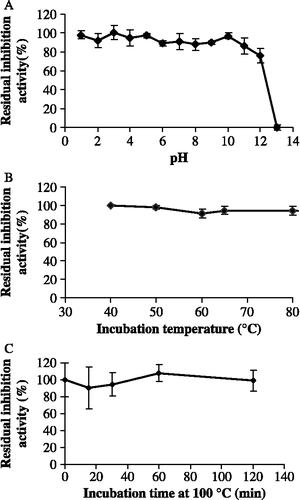
Figure 5. SDS-PAGE profile illustrating the resistance to proteolytic degradation by pepsin. TLXI samples (lanes 3 and 4) are compared with BSA samples (lanes 1 and 2) under reducing conditions, after 30 min at 37°C in the absence (lane 1 and 3) and the presence of pepsin (lane 2 and 4). The sizes of the Mr markers (lane 5) are indicated on the right. The gel was silver stained.
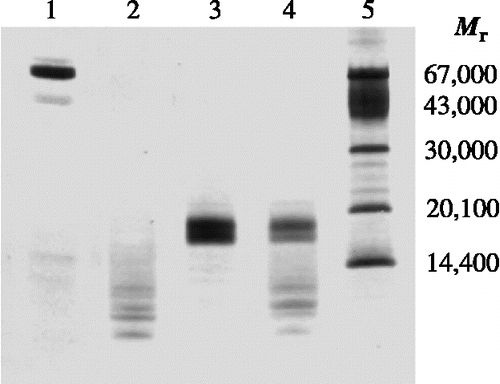
Figure 6. Relative inhibition activities of TLXI (▪) against XTL1 (A), XAN (B), XTV (C) and XPF (D) and relative xylanase activities in the absence of TLXI (♦) at different temperatures (left) and pH conditions (right).
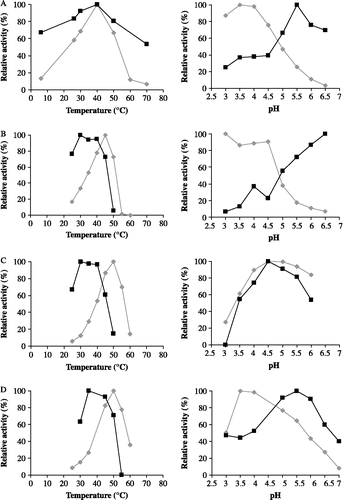
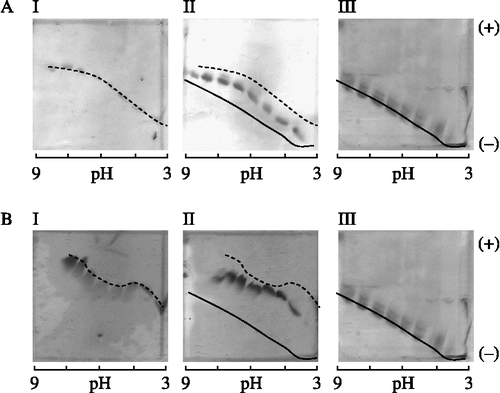
Figure 7. Iso-electric focusing titration curves showing the interaction between TLXI and XTL1 (A) and XAN (B). For each xylanase, a gel is shown with the xylanase (I), the mixture of xylanase and TLXI (II) and TLXI (III). The samples were incubated during 30 min at room temperature and pH 5.0 (25 mM sodium acetate buffer) prior to application on the gels. The cathode and anode are indicated by ( − ) and (+), respectively.