Figures & data
Figure 1. Typical gel filtration of Co-PTE and Zn-PTE. The size of Zn- or Co-PTE in solution was determined by gel filtration with an initial enzyme concentration of 1 mg/mL and a volume sample of 200 μL. A gel filtration of Co-PTE is shown. The absorbance profiles of Co-PTE (bold dashed line) and molecular weight standards (thin line) were recorded at 280 nm. The molecular weight of β-lactoglobulin (14.4 kDa), ovalbumin (43 kDa) and bovine serum albumin (67 kDa), and the estimated value of Co-PTE (36 kDa) are indicated at the top of each corresponding peaks.
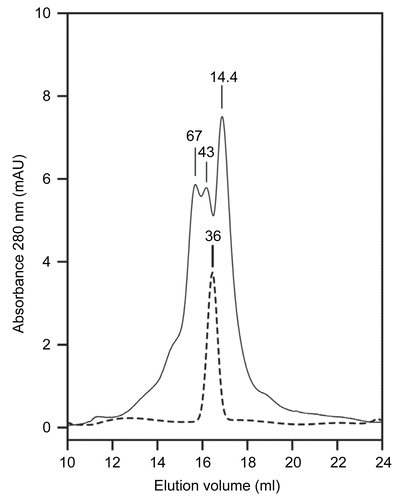
Figure 2. X-Ray Fluorescence spectrum of recombinant Zn-PTE and Co-PTE. XRF spectrum for energy between 5 and 10 KeV of 0.1 mM Zn-PTE (a) and 0.1 mM Co-PTE (b).
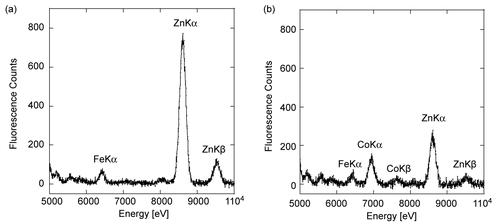
Figure 3. Stability of Zn-PTE and Co-PTE under assay conditions. Paraoxon hydrolysis of Zn-PTE and Co-PTE in the absence of additives, with 0.1% of β-lactoglobulin, with 50 μM ZnCl2 and with both additives, 5 minutes after dilution of the stock enzyme.
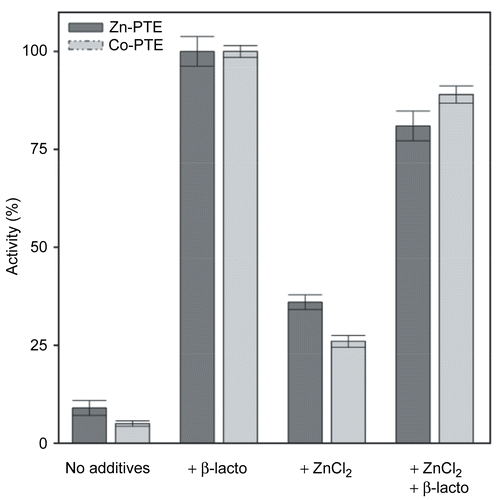
Figure 4. Michaelis-Menten Plot of paraoxon hydrolysis by Zn-PTE and Co-PTE. Initial reaction rate plotted against substrate concentration for paraoxon hydrolysis by PTE purified with Zn2+(○) and Co2+(□).
Table 1. Paraoxon hydrolysis catalyzed by recombinant wild type PTE expressed in the presence of zinc and cobalt chloride and mutants expressed in the presence of cobalt chloride.
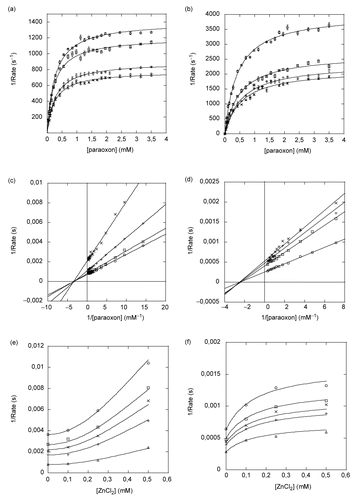
Figure 5. Inhibition of paraoxon hydrolysis of Co-PTE and Zn-PTE by ZnCl2. Reaction rate versus paraoxon concentration for Zn-PTE (a) and Co-PTE (b) in the absence (○) and presence of 0.1 mM (□), 0.25 mM (×) and 0.5 mM (+) ZnCl2. Lineweaver-Burk plot of Zn-PTE (c) and Co-PTE (d) activities in the absence (○) and presence of 0.1 mM (□), 0.25 mM (×) and 0.5 mM (+) ZnCl2. Dixon plots of Zn-PTE (e) and Co-PTE (f) with0.28 mM (○), 0.42 mM (□), 0.56 mM (×), 0.7mM (+), 3.5 mM (△) paraoxon. The best-fit curves or lines were determined using GOSA-fit and Equation (2), with n=2 for Zn-PTE and n=1 for Co-PTE.