Figures & data
Table 1. Cell wall sorting signals categorized by subfamily type.
Figure 1. The predicted quaternary structure of SaSrtA. The predicted His120, Cys184 and Arg197 in the active site are highlighted. The α-helix and β-sheet regions of the putative protein are indicated with cylinders and bands, respectively. Multiple sequence alignments revealed many conserved domains. The three amino acid residues thought to be involved in the formation of the catalytic site were conserved in those domains (highlights) of all 25 sequences. His120, Cys184 residues, participating in protein–protein interaction, are conserved between these bacteria above.
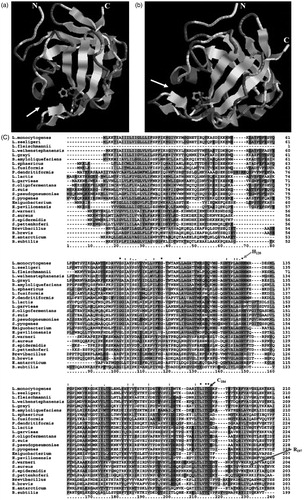
Figure 2. Phylogenetic relationship of the SrtA of various species. The SrtA used for phylogenetic tree construction are as the amino acid sequences used for homology alignment above. Numbers below the branches are the neighbor joining bootstrap values. The evolutionary distance is reflected by the numbers above the branches and the branch lengths proportional to the degree of amino acid substitutions. SrtAs exhibited a certain degree of homology among these nine groups.
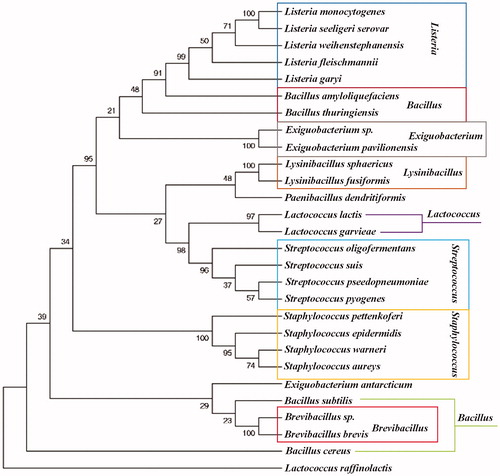
Figure 3. The structure of the LPXTG proteins precursor. (The length of rectangle does not stand for the real length of peptide). The LPXTG proteins, anchored by SrtA, mostly harbor an N-terminal signal peptide and a C-terminal cell wall sorting signal consisting of the LPXTG motif, hydrophobic domain and a positively charged tail have been found in all pathogenic Gram-positive bacteria, besides the anchored protein section.
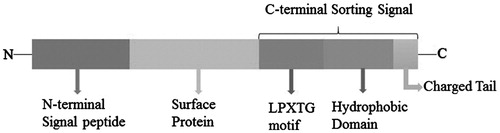
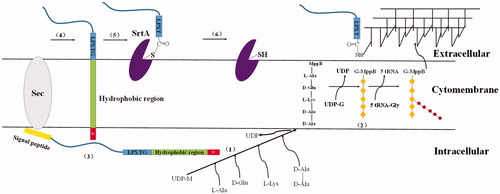
Figure 4. Cell wall sorting progress in Gram-positive bacteria. (1,2) Cell wall synthesis begins with the assembly of nucleotide linked wall peptides in the bacterial cytoplasm. Then the precursor, the lipid І, is transferred to bactoprenol pyrophosphate in the cytomembrane, next, N-Acetylglucosamine (G) connect to lipid І (MurNAc-(pentapeptide)-pyrophosphoryl-undecaprenol), forming the lipid II (GlcNAc-β-1,4-MurNAc-(pentapeptide)-pyrophosphoryl-undecaprenol). Finally lipid II molecules are transported across the cytoplasmic membrane for the polymerization reactions in bacteria. (3–6) Surface proteins are synthesized as precursors in the bacterial cytoplasm bearing an N-terminal signal peptide and a C-terminal sorting signal. The sorting signal contains an LPXTG sequence motif, a hydrophobic domain and a tail of positively charged residues. After surface protein synthesized, the N-terminal signal peptide will be removed before secreted out, and then the hydrophobic domain is embedded within the membrane and the charged tail remained in cell, retaining surface proteins in the secretory pathway. (5) The precursor cleaved by SrtA, a membrane-anchored transpeptidase, between threonines and glycine in the LPXTG motif, generating an acyl enzyme intermediate. (6) The thioester bond between SrtA and surface proteins, is resolved by the nucleophilic attack of the amino group of the cross-bridge (Gly5) within lipid II precursor. Finally, surface proteins linked to lipid II can be incorporated into the cell wall envelope.