Figures & data
Table 1. Pharmacophores generated from the co-crystallized Hsp90 structure (2XJX and 4LWF).
Table 2. Pharmacophoric features and corresponding weights, tolerances and 3D coordinates of generated pharmacophore from cocrystallized Hsp90 (2XJX_2_05).
Table 3. Pharmacophoric features and corresponding weights, tolerances and 3D coordinates of generated pharmacophore from cocrystallized Hsp90 (4LWF_2_04).
Table 4: NCI compounds with Hsp90 inhibitory values.
Figure 1. (A) 2XJX_2_05, the QSAR successful pharmacophore generated by DS studio 4.5, (B) cocrystallized XJX1232 in Hsp90 (2XJX, resolution 1.66 Å), (C) mapping of pharmacophore with cocrystallized Ligand XJX1232, (D) chemical structure of XJX1232.
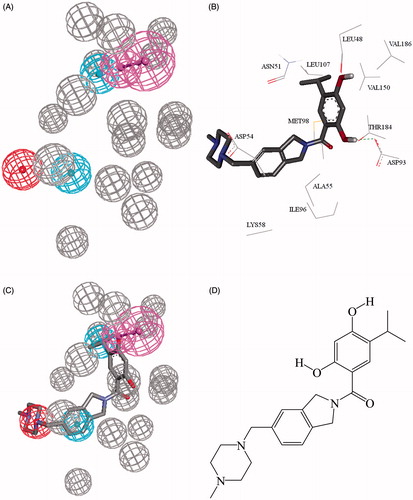
Figure 2. (A) 4LWF_2_04, the QSAR successful pharmacophore generated by DS studio 4.5, (B) cocrystallized FJ3301 in Hsp90 (4LWF, resolution 1.75 Å), (C) mapping of pharmacophore with cocrystallized Ligand FJ3301, (D) chemical structure of FJ3301.
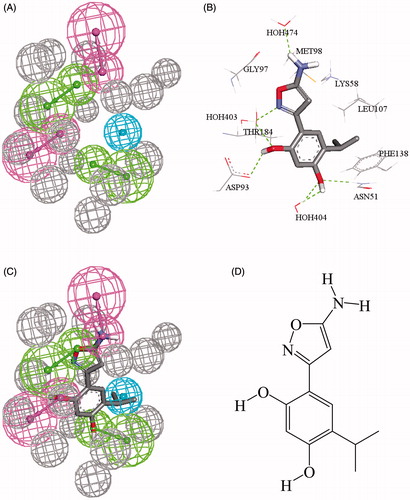
Figure 4. Two dimensional analysis of the cocrystallized ligand in the ATPase binding site of (A) Hsp90 (2XJX , resolution 1.66 Å) (B) Hsp90 (4LWF , resolution 1.75 Å), using DS 3.5 visualizer.
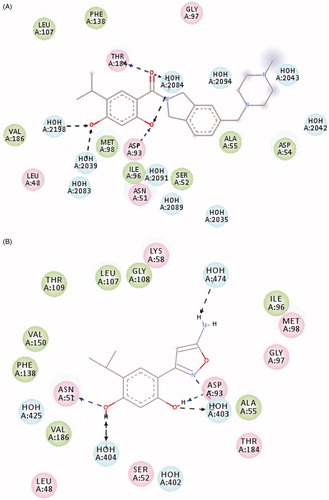
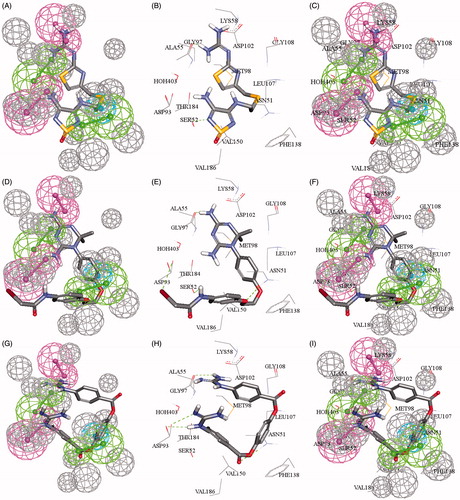
Figure 5. (A, D, G) Mapping compounds (84, 86, 88), respectively with 2XJX_2_05; cocrystallized pharmacophore generated by DS studio 4.5, (B, E, H) docked compounds (84, 86, 88), respectively in ATPase binding pocket of Hsp90 (2XJX, resolution 1.66 Å), (C, F, I) compounds (84, 86, 88) mapped and docked.
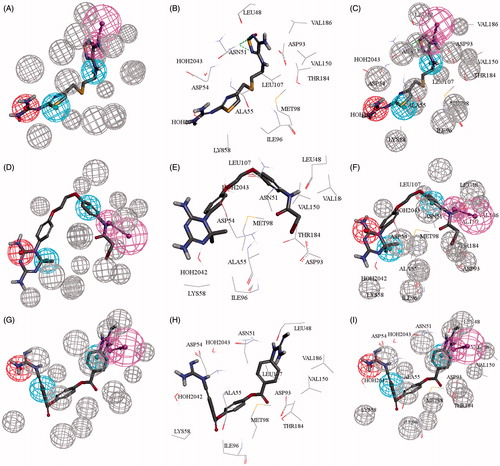
Figure 6. (A, D, G) Mapping compounds (84, 86, 88), respectively with 4LWF_2_04; cocrystallized pharmacophore generated by DS studio 4.5, (B, E, H) docked compounds (84, 86, 88), respectively in ATPase binding pocket of Hsp90 (4LWF, resolution 1.75 Å), (C, F, I) compounds (84, 86, 88) mapped and docked.