Figures & data
Table 1. α- and β-CA expressing nematoda, platyhelminthes, annelida, and hemichordate.
Figure 1. Multiple sequence alignment (MSA) of α-CA protein sequences from nematodes. MSA of 43 and one α-CA protein sequences from nematodes and Klebsiella pneumoniae (outgroup), respectively, aligned using Clustal Omega algorithm from EMBL-EBI database (http://www.ebi.ac.uk/Tools/msa/clustalo/)Citation45. MSA was conducted on 60 amino acids of α-CA protein sequences starting three amino acids prior to the first highly conserved histidine. The three highly conserved histidines locate in the catalytic active site of α-CAs and participate in binding with the Zn2+ ion (shown by red arrows).

Figure 2. Multiple sequence alignment (MSA) of β-CA protein sequences from nematodes. MSA of 31 and one β-CA protein sequences from nematodes and Bacillus subtilis (outgroup), respectively, aligned using Clustal Omega algorithm from EMBL-EBI database (http://www.ebi.ac.uk/Tools/msa/clustalo/)Citation45. MSA was conducted on 115 amino acids of β-CA protein sequences starting three amino acids prior to the first highly conserved sequence (CXDXR; C: Cysteine, D: Aspartic acid, R: Arginine, D: any residue). The first (CXDXR) and second (HXXC; H: Histidine, C: Cysteine, X: any residue) highly conserved sequences locate in the catalytic active site of β-CAs and are coordinated with the Zn2+ ion (shown by green and blue arrows, respectively).
Figure 3. Phylogenetic analysis of nematode α-CAs. A total of 54 nematode α-CAs were aligned using Clustal Omega and used to perform a phylogenetic analysis using the LG model in the PhyML program, with 1000 bootstraps. Node values indicate bootstrap values. Any sequences which were predicted from the full genomic sequence are labeled as “predicted”.
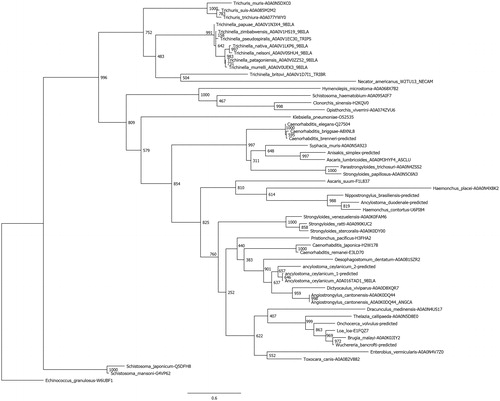
Figure 4. Phylogenetic analysis of nematode β-CAs. A total of 41 nematode β-CAs were aligned using Clustal Omega and used to perform a phylogenetic analysis using the LG model in the PhyML program, with 1000 bootstraps. Node values indicate bootstrap values. Any sequences which were predicted from the full genomic sequence are labeled as “predicted”.
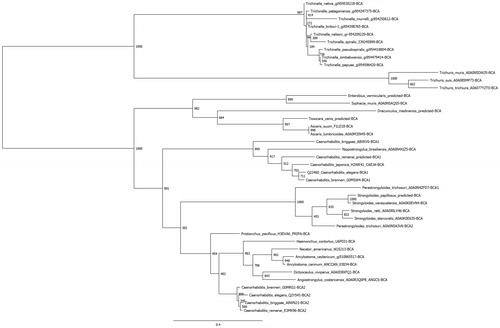
Table 2. Enzyme activity and inhibition data of β-CAs from protozoa, nematode, and insects.
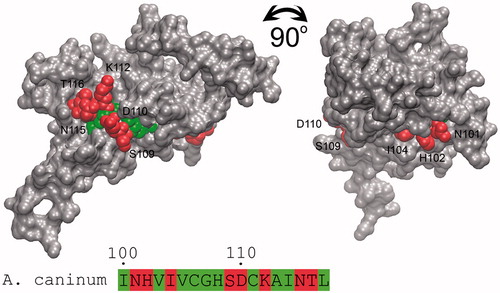
Figure 5. Accessibility identification of the predicted antigenic epitope of β-CA from A. caninum. The molecular surface of the homology model of A. caninum β-CA is shown as solid gray, and the second highly conserved sequence (HXXC) as the target epitope is buried from the surface of the protein. The exposed and buried residues of epitope are shown with red and green spheres and numbered. Figure adopted with author’s permission from open accessCitation10.