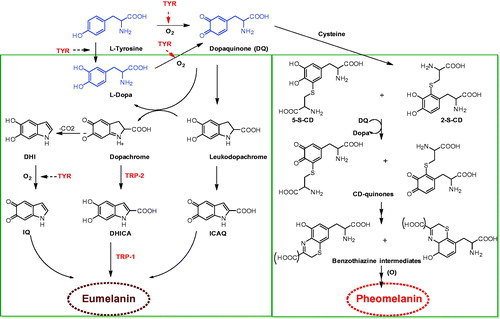
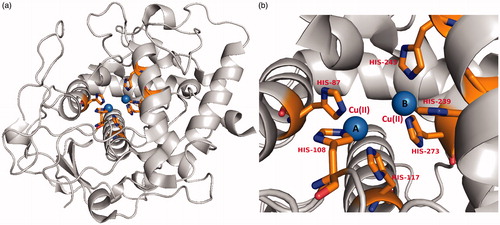
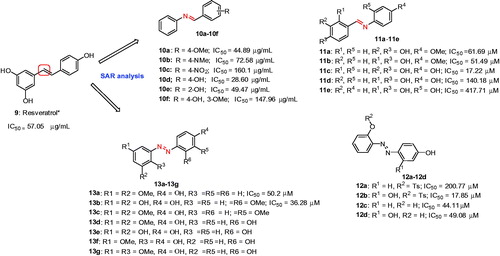
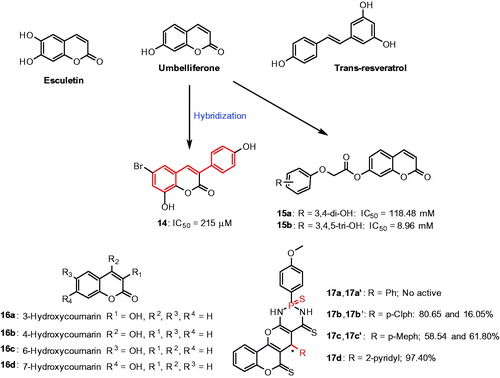
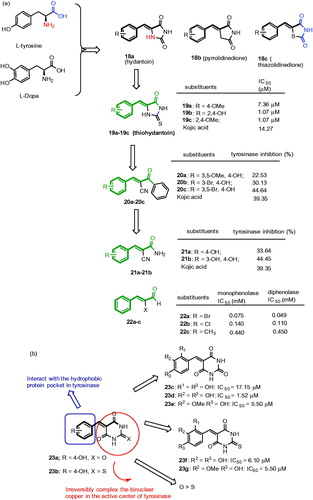
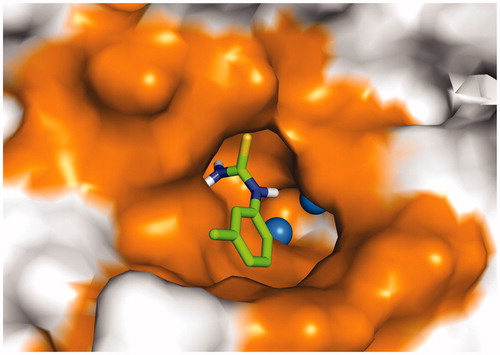
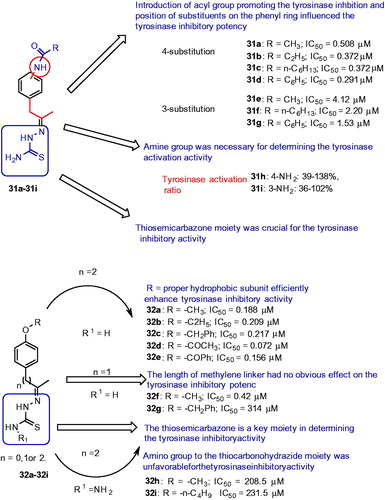
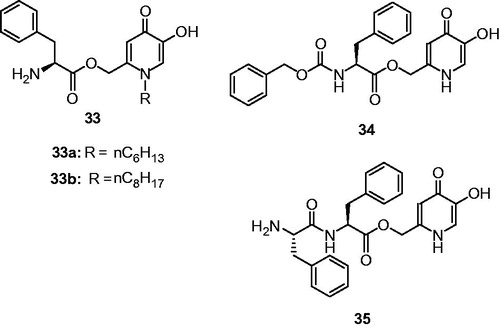
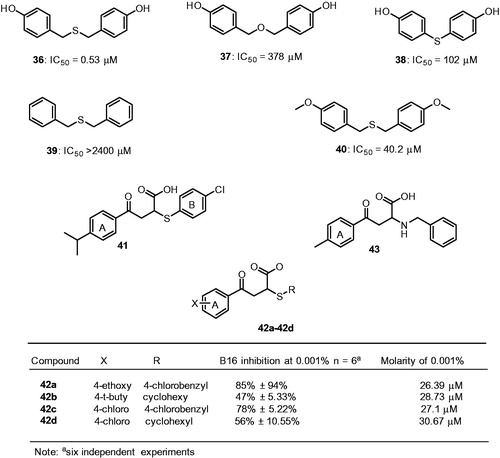
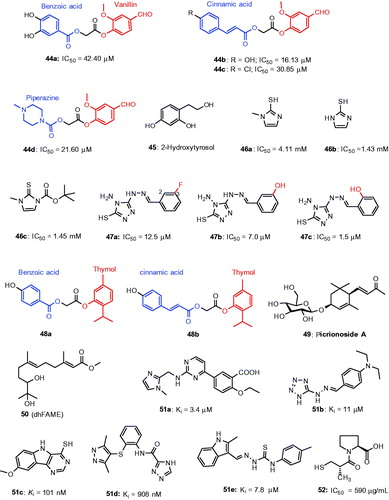
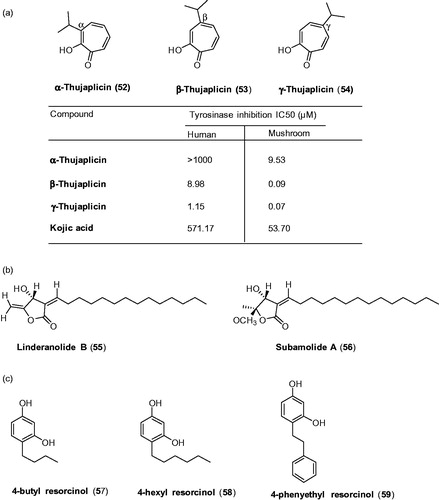
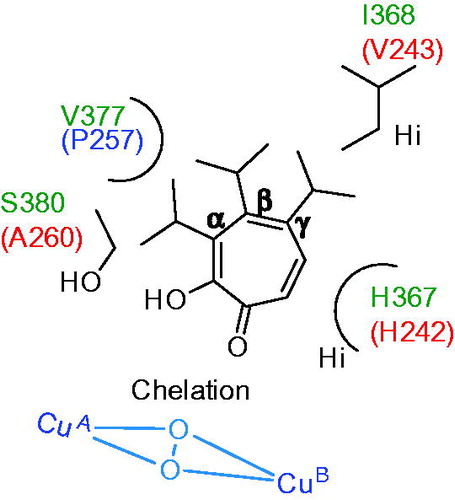
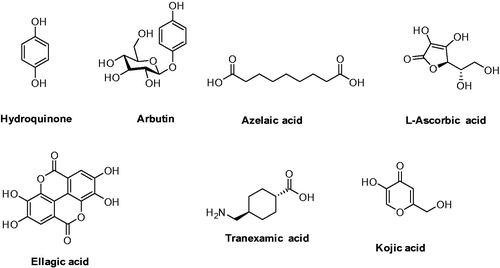
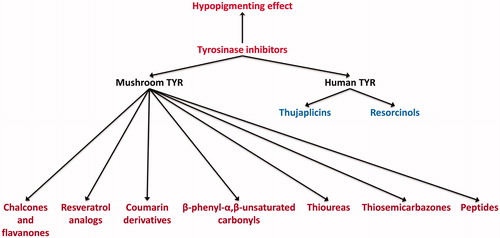
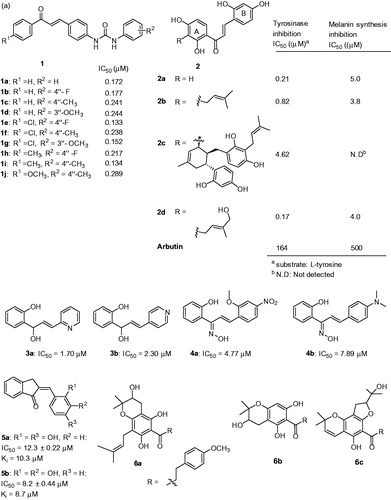
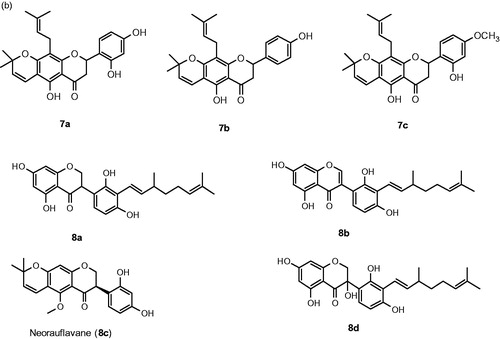
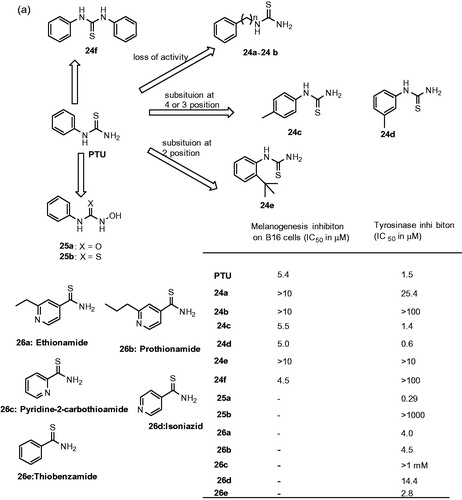
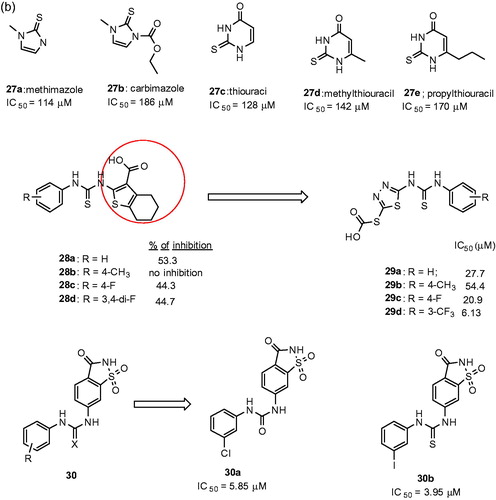
Figures & data
Table 1. Inhibition of melanin content in melanocytes by dipeptidesCitation111.