Figures & data
Figure 1. (A) Elution profile from the anion-exchange chromatography column of the supernatant obtained from the ammonium sulfate precipitation. Dot line represents the linear gradient from 0 to 0.5 NaCl; (B) Protonographic analysis carried out on the peaks eluted from the column. The yellow band denotes the hydratase activity due to the native CA purified by the mantles of the Mediterranean mussels, M. galloprovincialis.
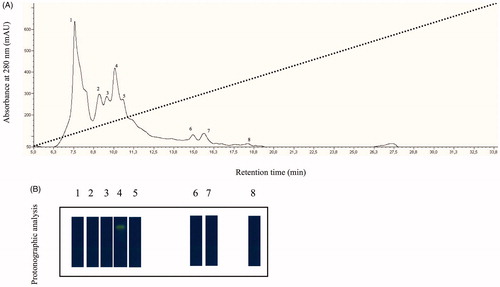
Figure 2. SDS-Page of the native CA purified from the mantles of M. galloprovincialis. Panel A, lane 2: cell extract protein from the mussel mantles before the purification; Panel B, lane 3: purified CA from pAMBS affinity column. Panel A and B, lane 1: molecular markers. Starting from the top: 250 kDa, 150 kDa, 100 kDa, 75 kDa, 50 kDa, 37 kDa, 25 kDa, 20 kDa, 15 kDa and 10 kDa.
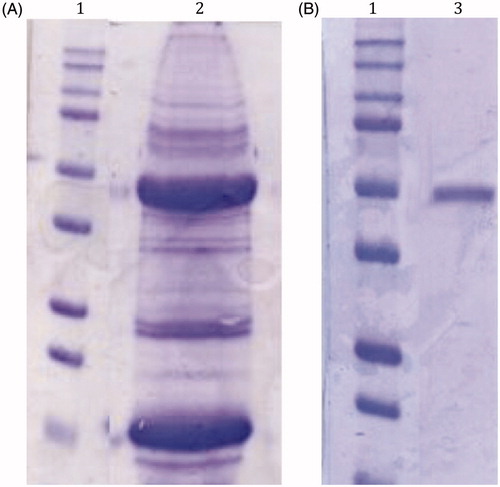
Figure 3. Protonogram obtained using the native CA from Mytilus galloprovincialis and the commercial bovine CA (bCA). Lane 2: mussel CA protonogram showing a band (yellow color) with an apparent molecular weight of 50 kDa; Lane 1: bovine CA is present in a monomeric state corresponding at a molecular weight of about 26 kDa; Lane 1: molecular markers. Starting from the top: 75 kDa, 50 kDa, 37 kDa, 25 kDa and 20 kDa.
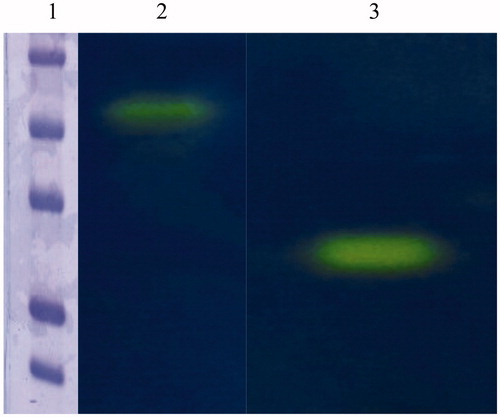
Figure 4. Amino acid sequence of CA from Mytilus galloprovincialis deposited in the protein data bank. In bold, the N-amino terminal sequence obtained from the electroblotted mussel CA. The asterisk indicates amino acid identity.

Figure 5. Amino acid multialignment obtained using the Mytilus galloprovincialis CA and Homo sapiens CA isoforms (hCA I and hCA II). The zinc ligands (His94, 96 and 119) and the gate-keeper residues (Glu106 and Thr199) are conserved in the mussel and mammalian sequences. The proton shuttle residue (His64) is missing in the M. galloprovincialis enzyme. hCA I numbering system was used. The asterisk (*) indicates identity at all aligned positions; the symbol (:) relates to conserved substitutions, while (.) means that semi-conserved substitutions are observed. Multialignment was performed with the program Muscle, version 3.1.

Table 1. Kinetic parameters for the CO2 hydration reaction catalyzed by the purified native mussel CA, the Homo sapiens CA isoforms (hCA I and hCA II) and coral CA isoforms (STPCA and STPCA-2). Acetazolamide (AAZ) inhibition data are also shown.
