Figures & data
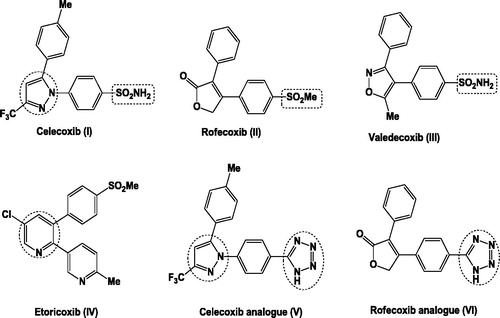
Table 1. In vitro COX-1 and COX-2 inhibition of tested compounds and reference drug, celecoxib.
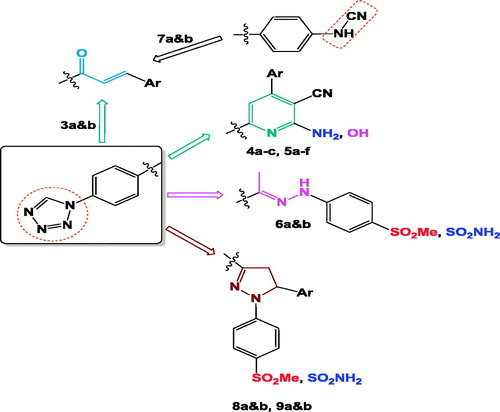
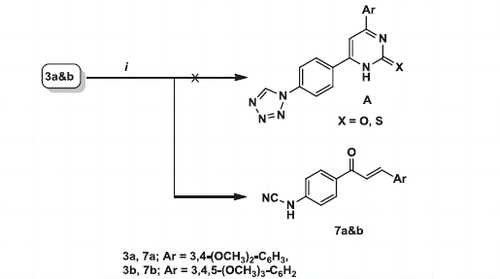
Scheme 1. Reagent and conditions: (i) NaN3, TEOF, gl. HAc, reflux, 12 h, (ii) ArCHO, KOH, abs. EtOH, r.t., 10–12 h, (iii) ArCHO, CN(CH2)CN, NH4OAc, abs. EtOH, (iv) ArCHO, CNCH2COOEt, NH4OAc, abs. EtOH, (v) CN(CH2)CN, NH4OAc, abs. EtOH, (vi) CNCH2COOEt, NH4OAc, abs. EtOH, (vii) p-substitutedphenylhydrazine hydrochloride, abs. EtOH, reflux, 6–8 h.
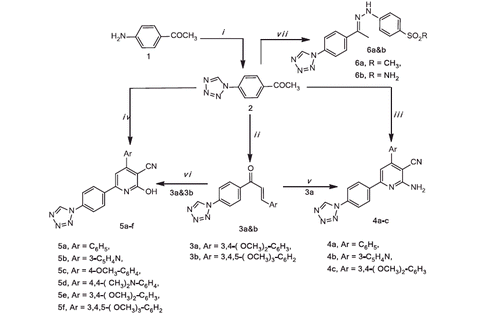
Scheme 3. Reagents and conditions: (i) p-methanesulphonylphenyl hydrazine hydrochloride, abs. ethanol; (ii) p-benzene sulphonamide hydrazine hydrochloride, abs. ethanol.
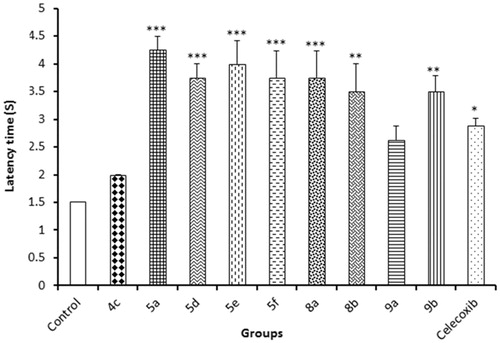
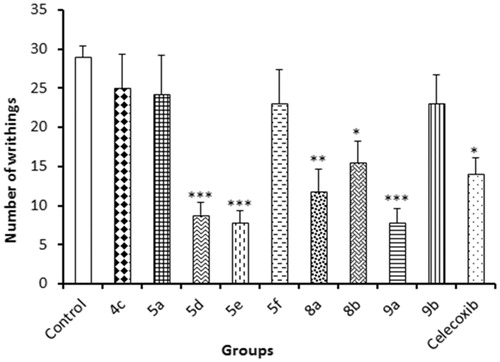
Table 2. Results of in vivo anti-inflammatory activities of tested compounds using carrageenan-induced rat paw oedema assay.
Table 3. Ulcerogenic liability for compounds 4c, 5a, 5d-f, 8a&b and 9a&b compared to reference drugs celecoxib and indomethacin.
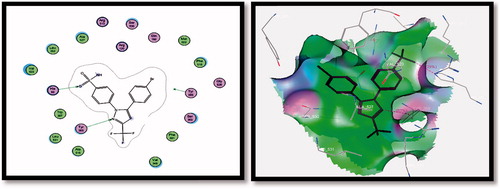
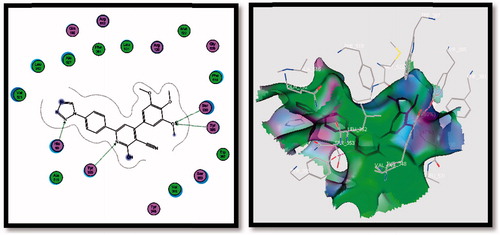
Table 4. Molecular modelling data for best poses of the designed compounds 3a&b, 4a-c, 5a-f, 6a&b, 7a&b, 8a&b, 9a&b and SC-558 during docking in COX-2 (PDB: 1CX2) active site.
Figure 5. Haematoxylin and eosin immunohistochemical staining of gastric ulcers after ulcer induction in rats for specimen intact Mucous membrane in control, indomethacin, celecoxib-treated rat and test compounds 4c, 5a, 5d and 5e.
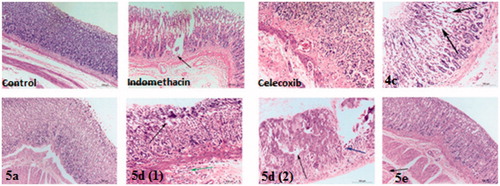
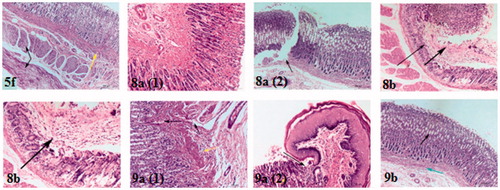
Figure 6. Haematoxylin and eosin immunohistochemical staining of gastric ulcers after ulcer induction in rats for specimen intact mucous membrane in 5f, 8a, 8 b, 9a and 9 b-treated rats.