Figures & data
Figure 1. Blast output reporting the CA library sequences. By going down the list, it is possible to see less than perfect matches, slowly degrading as the corresponding score decreases and the E-value increases. The E-value is an assessment of the statistical significance of the score. E-value close to 1 are a warning that the alignment is not reliable.
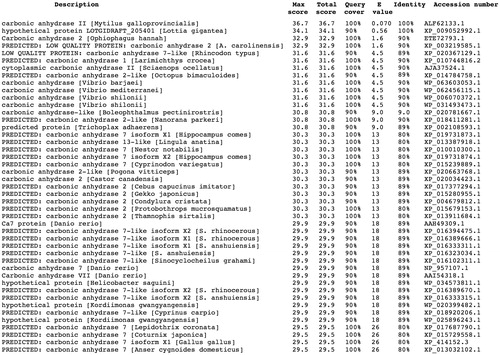
Figure 2. Alignment of α-CA sequences from Mytilus galloprovincialis (MgaCA, Accession number: ALF62133.1) Corallium rubrum (CruCA4, Accession number: KU557746), Homo sapiens I (hCAI, Accession number: NP001729) and II (hCAII, Accession number: P00918), hCA I numbering system was used. The zinc ligands (His94, 96 and 119) and the gate-keeper residues (Glu106 and Thr199) are conserved in aligned sequences; while the proton shuttle residue (His64) is preserved only in the human enzymes. The asterisk (*) indicates identity at all aligned positions; the symbol (:) relates to conserved substitutions, while (.) means that semi-conserved substitutions are observed. Multialignment was performed with the program Muscle, version 3.1.

Figure 3 (A and B). A. SDS-PAGE; B. Protonography. In both panels: Lane 1, Molecular weight markers; lane 2, the native MgaCA purified from the mantles of M. galloprovincialis; lane 3, the recombinant rec-MgaCA, lane 4, the commercial bovine CA.
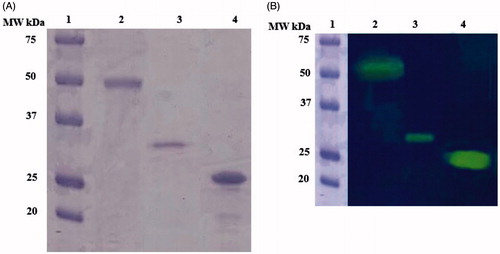
Table 1. Kinetic parameters for the CO2 hydration reaction catalysed by the rec-MgaCA, the purified native mussel CA (MgaCA), the Homo sapiens CA isoforms (hCA I and hCA II) and coral CA isoforms (SpiCA1 and SpiCA2 from Stylophora pistillata; CruCA4 from Corallium rubrum). Acetazolamide (AAZ) inhibition data are also shown.
