Figures & data
Table 1. Activity of Compounds 1–3 against DENV-2 Replication and DENV-2 NS5 RdRp.
Table 2. Activity of DENV inhibitors 4 and 5 against DENV-2 replication.
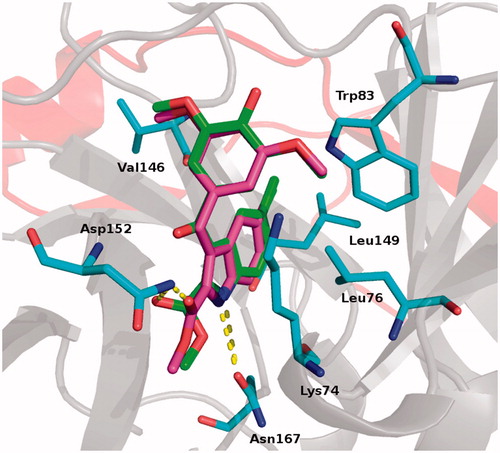
Figure 1. Proposed binding for derivatives 4 (green) and 5 (magenta). Residues involved in interactions are reported as cyan stick. H-bonds are shown as yellow dot lines. Protease is reported as cartoon: grey for NS3 and red for NS2 subunits.
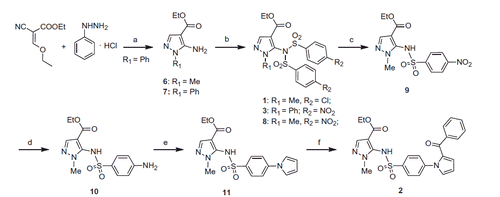
Scheme 1. Synthesis of the final compounds 1–3. (a) TEA, reflux, 7 h. (b) 4-Cl- or 4-NO2-C6H4SO2Cl, pyridine, 60 °C, overnight. (c) LiOH, 50 °C, 15 min. (d) SnCl2, 80 °C, 3 h. (e) 2,5-(OMe)2-THF, 100 °C, 1 h. (f) benzoyl chloride, AlCl3, closed vessel, 110 °C, 70 W, 4 min.
Scheme 2. Synthesis of the final compounds 4 and 5. (a) 4-OH-3,5-OMe2- or 3,5-OMe2-C6H4COCl, AlCl3, closed vessel, 110 °C, 70 W, 2 min. (b) TES, TFA, 48 h, rt.
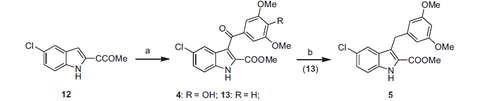
Figure 2. Inhibition of DENV RNA replication and RdRp activity by compounds 1–3. Panel A. Huh-7 cells were infected with DENV-2 at a multiplicity of infection (M.O.I) of 0.2 and followed by treatment with the DENV polymerase inhibitor for 3 days. The DENV RNA level was analysed by RT-qPCR with specific primer targeting viral NS5 gene, and relative viral RNA levels were normalised against cellular GADPH mRNA levels. Treatment of 50 μM 2′-C-methylcytidine (2CMC) direct against DENV RdRp served as positive control. 0.1% DMSO (Mock) served as negative control. Panel B. Huh-7 cells were transfected with pEG(MITA)SEAP and pcDNA-NS2B-GSG-NS3-Myc followed by incubation with each compound. The luciferase activity was analysed after 3 days treatment. Error bars denote the means ± SD of three independent experiments. *p < .05; **p < .01.
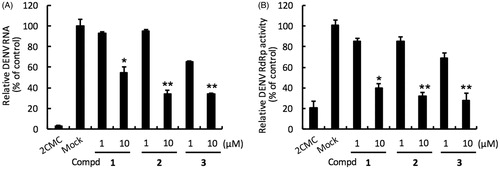
Figure. 3. Inhibition of DENV RNA and NS3-protease activity by compounds 4 and 5. Panel A. Huh-7 cells were infected with DENV-2 at a multiplicity of infection (M.O.I) of 0.2 and followed by the treatment of each DENV protease inhibitors for 3 days. The DENV RNA level was analysed by RT-qPCR with specific primer targeting viral NS5 gene, and relative viral RNA levels were normalised against cellular GADPH mRNA levels. Treatment of 50 μM 2′-C-methylcytidine (2CMC) direct against DENV RdRp served as positive control. 0.1% DMSO (Mock) served as negative control. Panel B. Huh-7 cells were transfected with pEG(MITA)SEAP and pcDNA-NS2B-GSG-NS3-Myc followed by incubation of each compounds, and the luciferase activity was analysed after 3 days treatment. Error bars denote the means ± SD of three independent experiments. *p < .05; **p < .01.
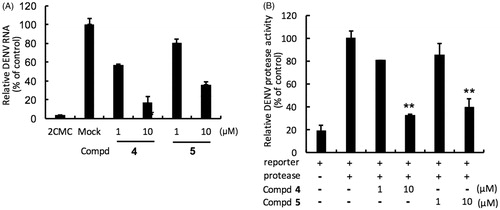
Figure 4. DENV RdRp inhibitor 3 protected ICR-suckling mice from DENV infection. 6-day-old ICR-suckling mice were intracerebrally injected with heat-inactive DENV (iDENV, n = 5) or active DENV (DENV, n = 4). Mice-receiving DENV were treated with 10 mg/kg of compound 3 (n = 5) at 1, 3, 5 dpi. Panel A, clinical scores; Panel B, body weight and Panel C, survival rate were recorded every day. Disease severity was scored as follow: 0: healthy, 1: slightly sick (reduced mobility), 2: inappetance, 3: weight loss and difficult to move, 4: paralysis, 5: death. Each group included 6 mice. Error bars denote the means ± SD.
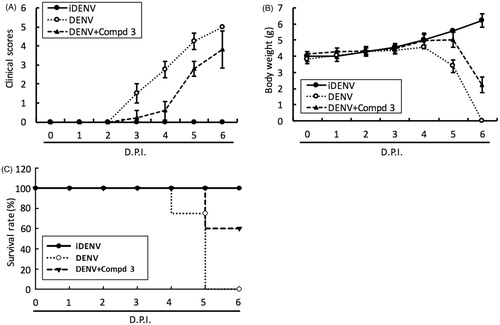
Figure 5. DENV protease inhibitor 4 protected ICR-suckling mice from DENV infection. Six-day-old ICR-suckling mice were intracerebrally injected with heat-inactive DENV (iDENV, n = 6) or active DENV (DENV, n = 6). Mice-receiving DENV were treated with 1 mg/kg of compound 4 (n = 6) at 1, 3, 5 dpi. Panel A, clinical scores, Panel B, body weight, and Panel C, survival rate were recorded every day. Disease severity was scored as follow: 0: healthy, 1: slightly sick (reduced mobility), 2: inappetance, 3: weight loss and difficult to move, 4: paralysis, 5: death. Each group included six mice. Error bars denote the means ± SD.
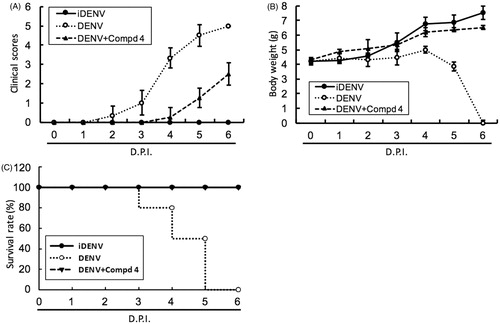
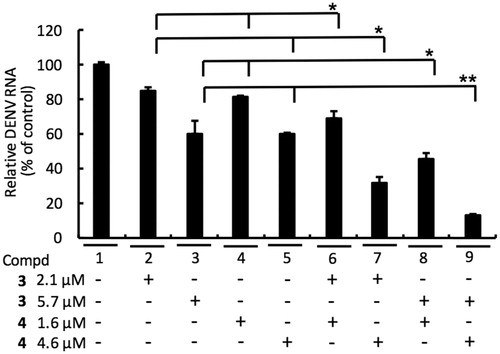
Figure 6. Combination assay of RdRp (3) and NS3 protease (4) inhibitors. Huh-7 cells were infected with DENV-2 at a multiplicity of infection (M.O.I) of 0.2 and followed by treatment of 3 and 4 with indicated concentration for 3 days. The DENV RNA level was analysed by RT-qPCR with specific primer targeting viral NS5 gene, and relative viral RNA levels were normalised against cellular GADPH mRNA levels. Error bars denote the means ± SD of three independent experiments. *p < .05; **p < .01.