Figures & data
Scheme 1. Reagents and conditions: (i) DCC, DMAP, DCM, substituted aromatic phenolic compounds, room temperature, 2 h.
Table 1. Chemical structures of the title compounds.
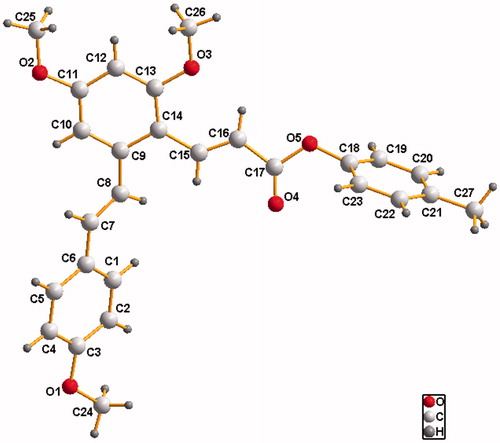
Table 2. Crystallographic data and structure refinements for compound D2.
Table 3. Selected bond lengths (Å) and angles (°) for compound D2.
Figure 3. Effects of compounds D1–D23 on production of NO by RAW264.7 cell RAW264.7 cells were pretreated with D1–D23 (40 μM) for 4 h, and then stimulated with or without LPS (1 μg/mL) for 24 h. NO production was measured using nitrite and nitrate assay. ###p < .001 compared with unstimulated cells, *p < .05, **p < .01 and ***p < .001 compared with LPS-stimulated cells; Data were from at least three independent experiments, each performed in duplicate.
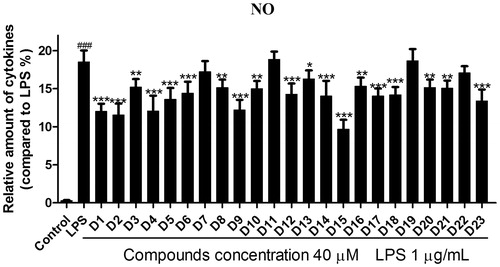
Figure 4. Compound D15 inhibited LPS-induced inflammatory response in RAW 264.7 cells. Cells were treated with compound D15 (10, 20, 40 µM) for 12 h, and then stimulated by LPS (1 µg/ml) for 3 h. Cell viability was evaluated using the MTT assay. NO production was measured using nitrite and nitrate assay. iNOS and COX-2 expression were detected by Western blot analysis. (A) Cell viability assay; (B) Quantitative analysis of NO production. (C) Quantitative analysis of iNOS and COX-2 expression, β-actin was used as loading control. ###p < .001 compared with unstimulated cells, **p < .01 and ***p < .001 compared with LPS-stimulated cells. Data were from at least three independent experiments, each performed in duplicate.
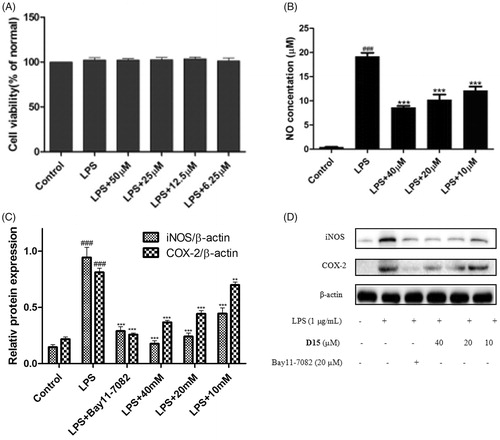
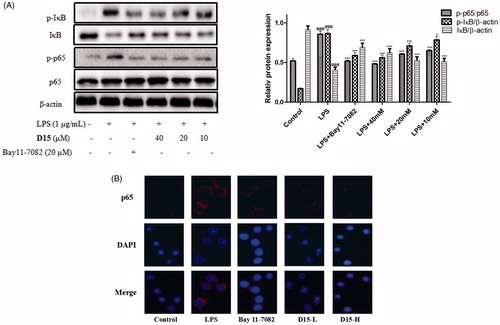
Figure 5. Compound D15 suppressed LPS-induced activation of NF-κB signaling pathway in RAW 264.7 cells. After pretreatment with D15 (10 ∼ 40 µM), RAW 264.7 cells were stimulated with LPS (1 µg/mL) for 30 min. The total and phosphorylation levels of NF-κB were detected by Western blot. (A) Quantitative analysis of p-IκB and p-p65, total IκB and p65 were used as loading control, respectively. (B) Immunofluorescence analysis of compound D15. ###p < .001 compared with unstimulated cells, *p < .05 and ***p < .001 compared with LPS-stimulated cells. Data were from at least three independent experiments, each performed in duplicate.