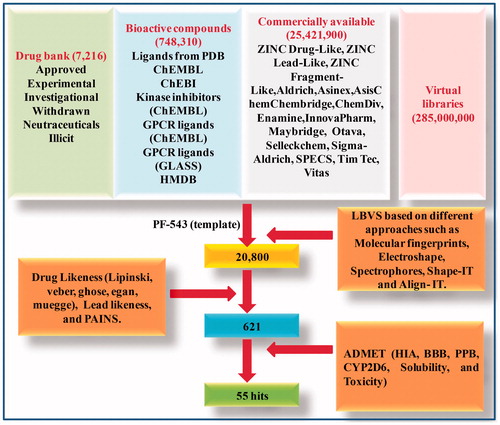
Figures & data
Table 1. Analysis of drug like properties of top 10 compounds and PF-543 on the basis of Lipinski’s RO5.
Table 2. ADMET calculation of top 10 compounds and PF-543.
Table 3. Docking analysis of top 10 compounds and comparison with known drug.
Figure 2. The binding mode of ZINC06823429 with the SphK1. (A) The overall structure of SphK1–ZINC06823429 complex showing protein in cartoon model and ligand in stick. (B) Interaction of ZINC06823429 to the SphK1 residues (stick). (C) 2D diagram of SphK1 interaction with the compound ZINC06823429. The active site residues of SphK1 interacting with ligand ZINC06823429 by four conventional H-bonds.
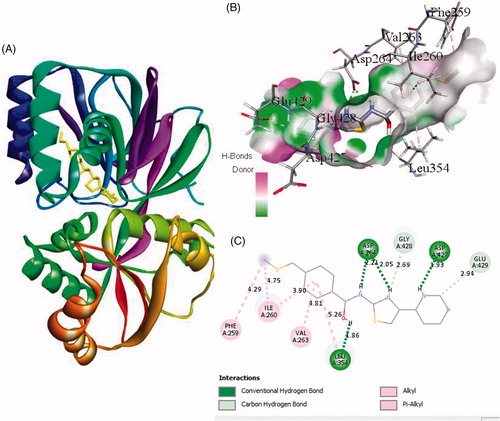
Figure 3. The binding mode of ZINC95421501 with the SphK1. (A) The overall structure of SphK1–ZINC95421501 complex showing protein in cartoon model and ligand in stick. (B) Interaction of ZINC95421501 to the SphK1 residues (stick). (C) 2D diagram of SphK1 interaction with the compound ZINC95421501. The active site residues of SphK1 interacting with ligand ZINC95421501 by two conventional H-bonds.
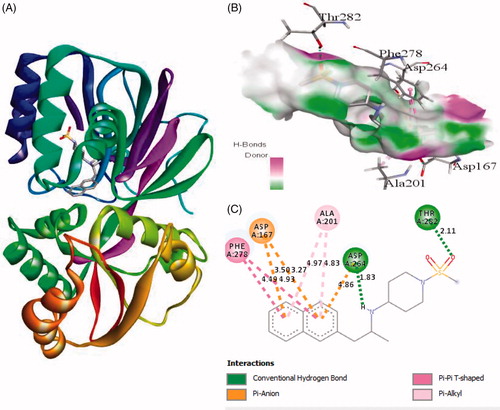
Figure 4. The binding mode of ZINC95421070 with the SphK1. (A) The overall structure of SphK1–ZINC95421070 complex showing protein in cartoon model and ligand in stick. (B) Interaction of ZINC95421070 to the SphK1 residues (stick). (C) 2D diagram of SphK1 interaction with the compound ZINC95421070. The active site residues of SphK1 interacting with ligand ZINC95421070 by three conventional H-bonds.
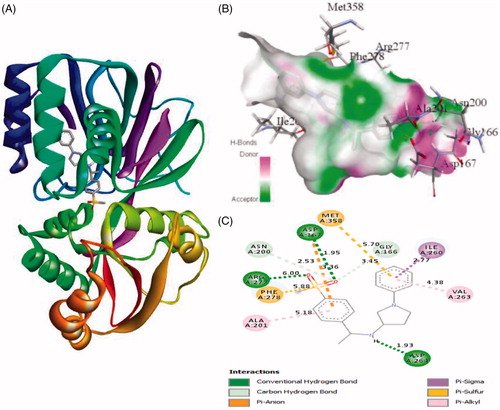
Figure 5. The binding mode of PF-543 with the SphK1. (A) The overall structure of SphK1–PF-543 complex showing protein in cartoon model and ligand in stick. (B) Interaction of PF-543 to the SphK1 residues (stick). (C) 2D diagram of SphK1 interaction with the compound PF-543. The active site residues of SphK1 interacting with ligand PF-543 by one conventional H-bonds.
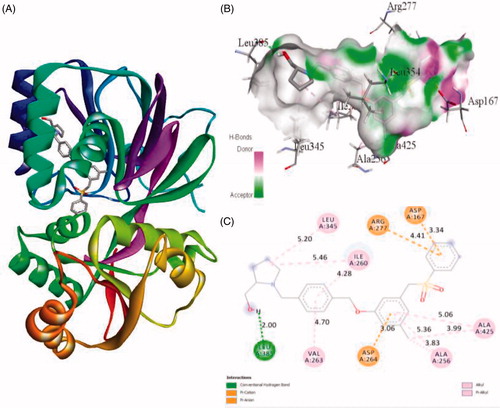
Table 4. Calculated parameters for all the systems obtained after 100 ns MD simulations.
Figure 6. Dynamics of ligands binding to the SphK1. (A) RMSD plot as a function of time. Black, red, green, blue, and yellow colours represent values obtained for SphK1, SphK1–STD, SphK1–ZINC06823429, SphK1–ZINC95421070, and SphK1–ZINC95421501, respectively. (B) Comparison of orientations of PF-543 (red), ZINC06823429 (green), ZINC95421070 (blue), and ZINC95421501 (yellow) into the active pocket of SphK1. (C) Backbone atomic fluctuations (RMSF) plot for SphK1, SphK1–PF-543, SphK1–ZINC06823429, SphK1–ZINC95421070, and SphK1–ZINC95421501. (D) Time evolution of radius of gyration (Rg) values during 100,000 ps (100 ns) of MD simulation. The Rg plot for SphK1 is shown in black. Red, green, blue, and yellow colours represent Rg plot for SphK1–PF-543, SphK1–ZINC06823429, SphK1–ZINC95421070, and SphK1–ZINC95421501, respectively.
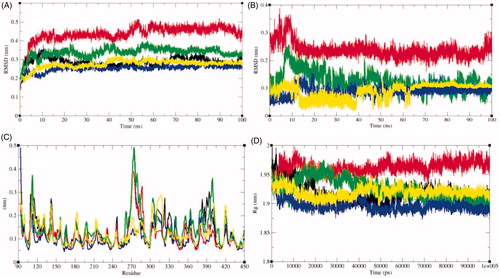
Figure 7. Solvent accessible surface area. (A) The solvent accessible surface area (SASA) as a function of time. Black, red, green, blue, and yellow colours represent values obtained for SphK1, SphK1–STD, SphK1–ZINC06823429, SphK1–ZINC95421070, and SphK1–ZINC95421501, respectively. The SASA plot was further resolved into hydrophobic and hydrophilic surface area for (B) SphK1, (C) SphK1–PF-543, (D) SphK1–ZINC06823429, (E) SphK1–ZINC95421070, and (F) SphK1–ZINC95421501, respectively.
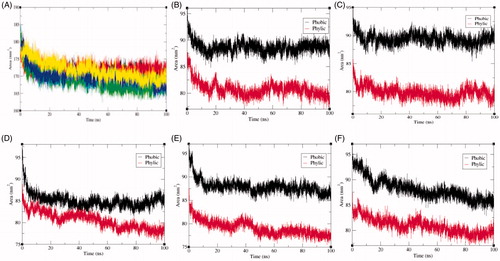
Figure 8. Free energy of solvation during SASA analysis. Free energy of solvation during SASA analysis was calculated for SphK1, SphK1–PF-543, SphK1–ZINC06823429, SphK1–ZINC95421070, and SphK1–ZINC95421501, respectively. Black, red, green, blue, and yellow colours represent the values obtained for SphK1, SphK1–PF-543, SphK1–ZINC06823429, SphK1–ZINC95421070, and SphK1–ZINC95421501, respectively.
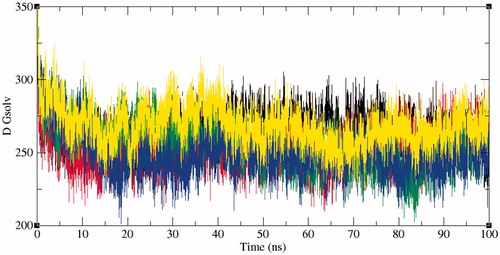
Figure 9. The average number of hydrogen bonds as a function of time. Red, green, blue, and yellow colours represent the number of hydrogen bonds for SphK1–PF-543, SphK1–ZINC06823429, SphK1–ZINC95421070, and SphK1–ZINC95421501, respectively.
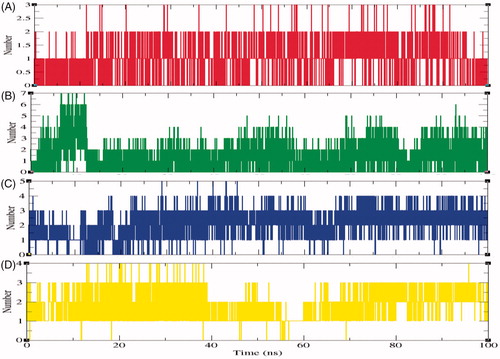
Figure 10. The secondary structure plot. The graphical representation indicating the structural elements present during 100 ns MD simulations in (A) SphK1, (B) SphK1–PF-543, (C) SphK1–ZINC06823429, (D) SphK1–ZINC95421070, and (E) SphK1–ZINC95421501, respectively.
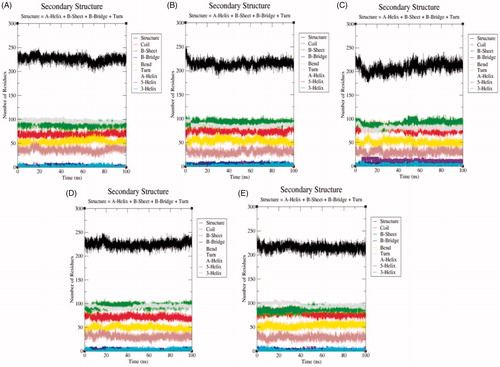
Figure 11. Position of ligands in the secondary structure framework. The secondary structure representation of SphK1 during 100 ns MD simulation in (A) SphK1, (B) SphK1–PF-543, (C) SphK1–ZINC06823429, (D) SphK1–ZINC95421070, and (E) SphK1–ZINC95421501, respectively.
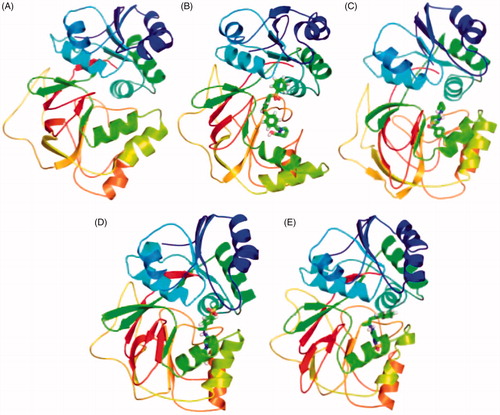
Table 5. Percentage of residues participated in average structure formation.
Figure 12. Principal component analysis (PCA). The PCA or essential dynamics (ED) calculated for SphK1, SphK1–PF-543, SphK1–ZINC06823429, SphK1–ZINC95421070, and SphK1–ZINC95421501, respectively. (A) The graph indicated the projection of eigenvectors in SphK1 (black), SphK1–PF-543 (red), SphK1–ZINC06823429 (green), SphK1–ZINC95421070 (blue), and SphK1–ZINC95421501 (yellow), respectively. (B) The Eigen root mean square fluctuations indicating the atomic fluctuations were also calculated for SphK1 (black), SphK1–PF-543 (red), SphK1–ZINC06823429 (green), SphK1–ZINC95421070 (blue), and SphK1–ZINC95421501 (yellow), respectively.
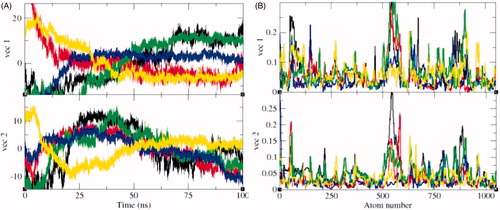
Figure 13. 2D projection of trajectories. The 2D projections of trajectories on eigenvectors showed different projections of SphK1 in case of (A) SphK1, (B) SphK1–PF-543, (C) SphK1–ZINC06823429, (D) SphK1–ZINC95421070, and (E) SphK1–ZINC95421501, respectively.
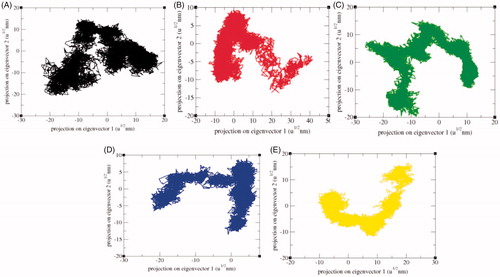
Table 6. Average binding energy calculation using g_mmpbsa package implemented in GROMACS for high-throughput MMPBSA calculation for protein–ligand complexes.