Figures & data
Table 1. In vitro enzymatic and antiparasitic activities of selected hGSK-3β inhibitors.Table Footnotea
Figure 1. Binding mode into the LmjGSK-3 enzyme for ITDZ, TDZD, and HMK representative compounds. (A) Blind docking poses obtained from the most representative clusters for 2 (TDZD) and 4 (ITDZ) in the ATP binding site and the substrate binding site, respectively. (B) Superimposition of most representative regular docking results of ITDZ compounds 3 and 4. (C) Superimposition of the best covalent docking poses obtained for TDZDs (1 and 2) and HMK 5. (D) Detailed view of the covalent docking for compound 2.
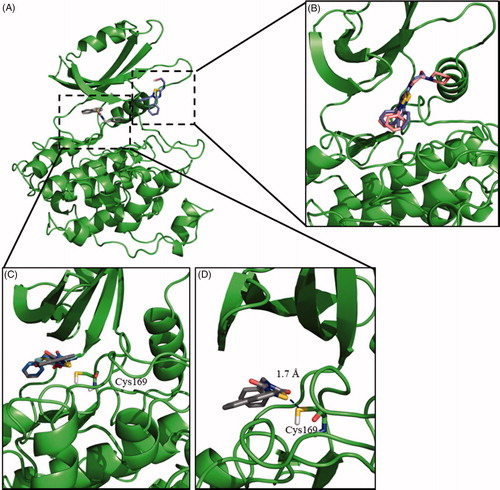
Figure 2. (A) Superposition of the human (purple) and Leishmania (green) GSK-3 enzymes. Key mutation in the ATP binding site is depicted as sticks. (B) Superposition of the poses of maleimide 13 in the human (purple) and Leishmania (green) GSK-3 enzymes.
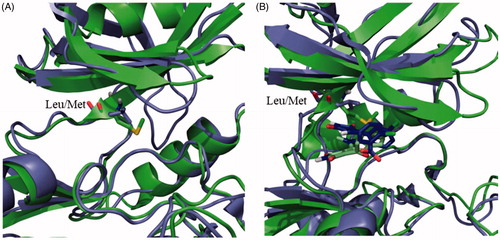
Table 2. IC50 values of hGSK-3 and LdGSK-3 inhibition for hit compounds 71, 95, 119, 124, 128, 151 and 187.
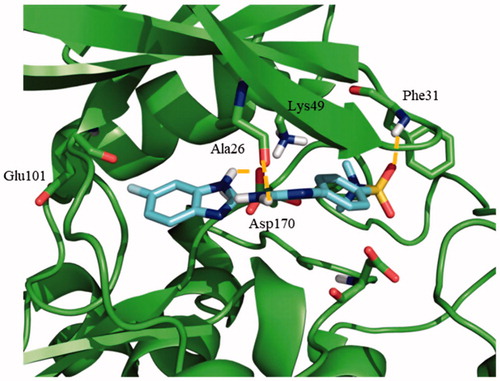
Figure 3. Proposed binding mode for compound 95 with LmjGSK-3 (PDB code 3E3P) with the modelled decapeptide loop. Zoom of ATP binding pocket showed the most relevant interactions of 95 with nearby residues of the binding site.
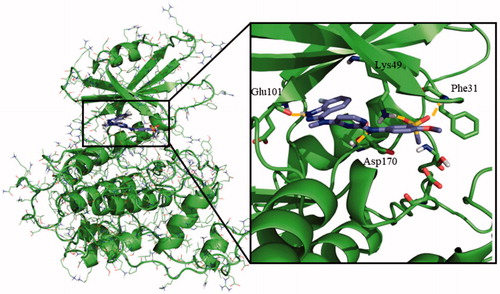
Figure 4. Docking results for the N-phenylpyrimidine-2-amines into modelled LmjGSK-3. (A) Superposition of compounds 124 (magenta), 119 (yellow) and 95 (purple) on LmjGSK-3 protein. Key residues were labelled. (B) Binding mode of compound 124. The main interactions were highlighted. (C) Binding mode of compound 119 showing the main interactions found in the complex.
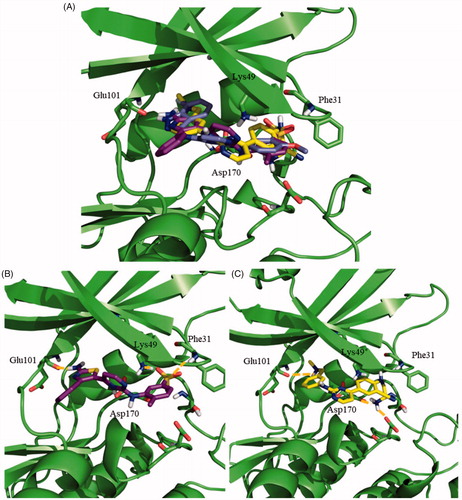
Figure 5. Proposed binding mode for the active benzoimidazole 71 into modelled LmjGSK-3 showing the main interactions found in the complex.