Figures & data
Table 1. Structure, inhibitory activity (IC50), antibacterial activity (MIC50) against H. pylori urease of compounds b1–b29.
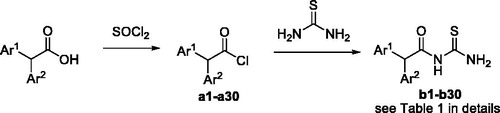
Table 2. The binding affinity data for urease–thioureas interactions.
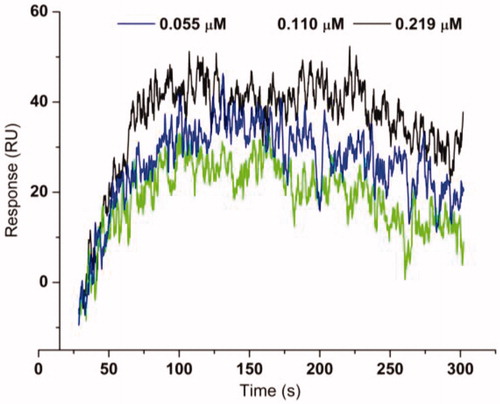
Figure 3. A velocity (V) was nonlinearly fitted against the concentrations of urea [S] in the presence of a specific concentration of compound b19. (B) (1) the fitted constants (
) from the corresponding V–S plot were plotted against concentrations of b19 ([I]); (2)
the fitted constants (
) from the corresponding V–S plot were plotted against concentrations of b19.
![Figure 3. A velocity (V) was nonlinearly fitted against the concentrations of urea [S] in the presence of a specific concentration of compound b19. (B) (1) Ki: the fitted constants (KmVmax(1+[I]Ki)) from the corresponding V–S plot were plotted against concentrations of b19 ([I]); (2) Ki': the fitted constants (1Vmax(1+[I]Ki′)) from the corresponding V–S plot were plotted against concentrations of b19.](/cms/asset/cbdcdec4-28e7-4e50-bece-e902474d1941/ienz_a_1706503_f0003_c.jpg)
Table 3. Data of inhibition mechanism.
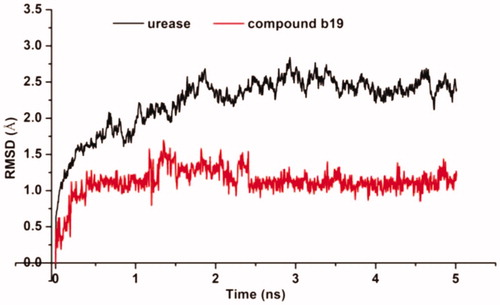
Figure 4. Predicted binding mode of ligand-urease (PDB code: 1e9y): (A) compound b19 shown as white sticks and the enzyme shown as surface. (B) Compound b19 shown as cyan sticks and enzyme shown as lines; Hydrogen bonds shown as red dashed lines.
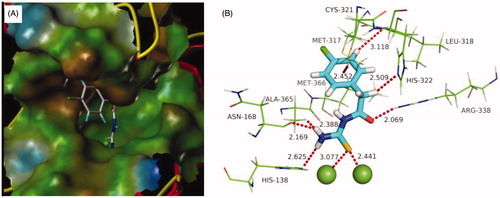
Table 4. Cell Viability of selected compounds on L-02 and P69 at concentration of 25 μg/mL.