Figures & data
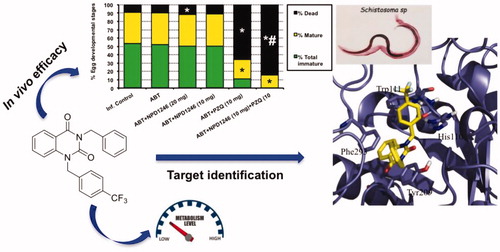
Figure 1. In vitro findings for NPD-1246 (1) previously reportedCitation14.

Table 1. In vitro metabolic stability of NPD-1246: percentage of parent compound remaining over time in the presence of mouse liver microsomes.
Figure 2. Effect of NPD-1246 alone (in a dose of 20 or 10 mg/kg/day) or in combination with PZQ (10 mg/kg/day each) for 5 days treatment on (A) worm burden, (B) tissue egg load and (C) oogram pattern in S. mansoni-infected mice sacrificed 10 days post end of treatment. *Significantly different from infected control at p < 0.05. #Significantly different from PZQ group at p < 0.05. Numbers above columns and between parentheses represent percentage change from infected control group.
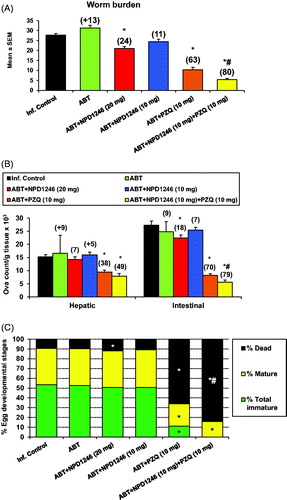
Figure 3. Metabolic site prediction using SMARTCyp web server for NPD-1246. Top-ranked sites (red circles) and minor sites (blue circles) predicted to be metabolised by (A) CYP2C9, (B) CYP2D6 and (C) CYP3A4.
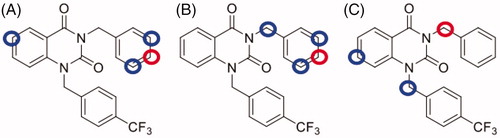
Table 2. Mature worm killing and ovipositing at 100 µM and 50 µM of new quinazolines (2–13) in comparison with previously reported data for NPD-1246 (1)Citation14.
Table 3. Mature (6 weeks old) worm killing and ovipositing under different concentrations of selected compounds in comparison with NPD-1246 (1) dataCitation14 previously reported.
Table 4. Mature (male & female) worm killing under different concentrations of selected compounds in comparison with NPD-1246 (1) dataCitation14 previously reported.
Table 5. In vitro metabolic stability of 9 and 10: percentage of parent compound remaining over time in the presence of mouse and human liver microsomes.
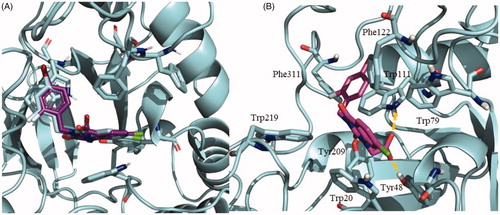
Figure 4. Quinazoline-related structures crystallised with different target proteins according to the scaffold search in the PDB (access codes included).
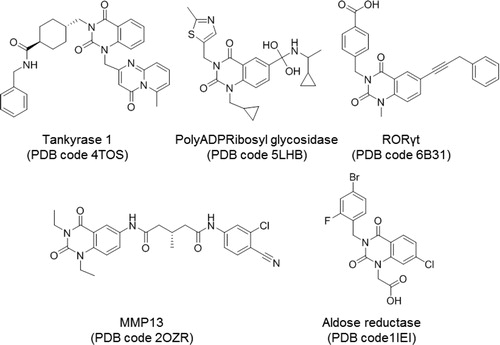
Figure 5. Superposition of the crystal structures of human aldose reductase 1IEI, depicted in cyan, S. japonicum aldose reductase 4HBK in magenta and the homology model of S. mansoni aldose reductase in purple, (A) front view and (B) back view.
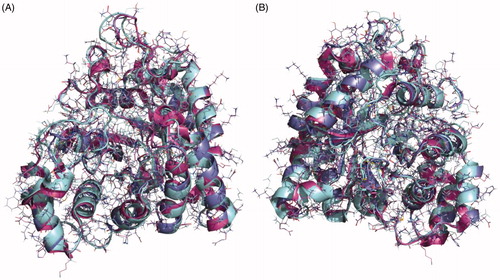
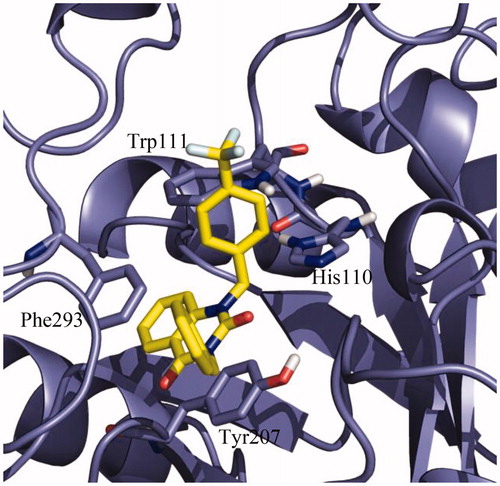
Figure 6. (A) Superimposition of zenarestat in the crystal structure 1IEI depicted in cyan and validation docking results depicted in purple show the high similarity between both poses in the human enzyme. (B) Detail of the zenarestat binding mode together with the main interactions found in the catalytic site of aldose reductase.