Figures & data
Figure 1. Amino acid sequence alignment of tyrosinases from Homo sapiens, Mus musculus, Oryzias latipes, Streptomyces antibioticus, Streptomyces glaucescens, Streptomyces lavendulae, Streptomyces lincolnensis, Neurospora crassa, Aspergillus oryzae, and Sinorhizobium meliloti. The numbers correspond to human tyrosinase amino acid positions, and copper ligands in human tyrosinase are indicated.

Table 1. Primers used for recloning into pET26b(+) and site-directed mutagenesis.
Figure 2. (A) SDS-PAGE of wild-type human tyrosinase and mutant enzymes after purification through diethylaminoethyl (DEAE)-Sephacel and immobilised metal-affinity chromatography. The gel was stained with Coomassie blue. The arrow indicates the calculated size of 66 kDa, corresponding to human tyrosinase. (B) Confirmation of tyrosinase stability by refolding. The tyrosinase expressed and purified in E. coli BL21 was denatured by adding 8 M urea, and then refolded by gradationally reducing the urea concentration by dialysis. The activity of the refolded tyrosinase was measured and compared to that of the early purified tyrosinase. The values represent the mean ± SD (n = 3).
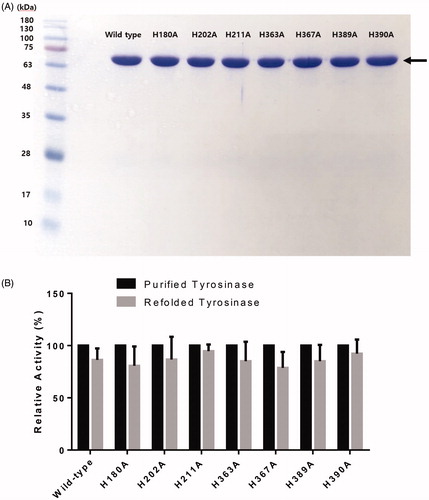
Figure 3. (A) Proposed three-dimensional structure of human tyrosinase. The di-copper site was found in the centre of the four-helix bundle motif. H180, H202 and H211 residues show direct binding with the copper atom at the CuA site, and H363, H367 and H390 are directly coordinated with the copper atom at the CuB site. However, H389 is located around the CuB site without direct binding. (B) Schematic illustration of CuA- and CuB-binding sites of human tyrosinase. Based on three-dimensional structure modelling, the residues H180, H202, and H211 show direct binding with CuA, whereas H363, H367 and H390 are directly coordinated with CuB. H389 is located around CuB but without direct binding.
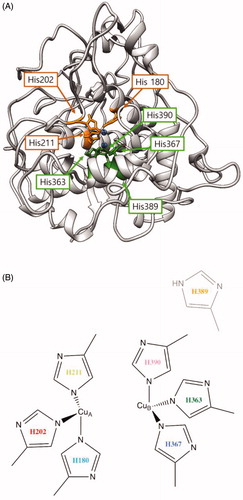
Table 2. Specific activity of the wild-type tyrosinase and mutant enzymes for l-tyrosine hydroxylation and l-dopa oxidation reactions.
Table 3. Kinetic parameters of the wild-type tyrosinase and mutant enzymes for l-tyrosine hydroxylation and l-dopa oxidation reactions.
