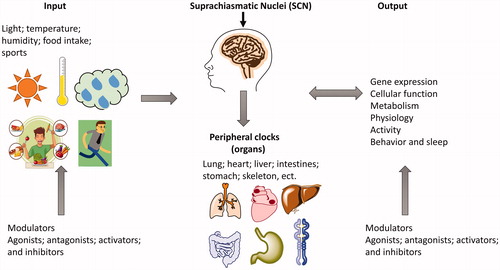
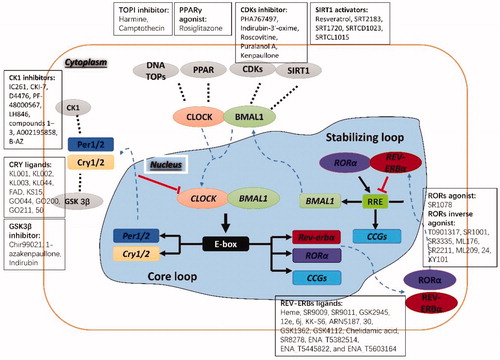
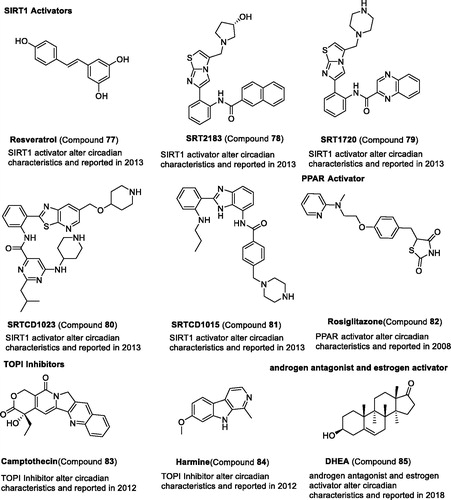
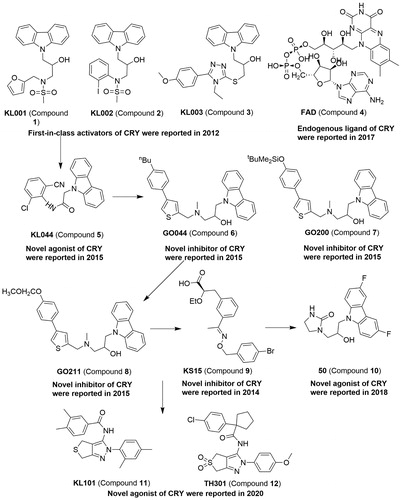
Figures & data
Table 1. Modulators targeting CRYs.
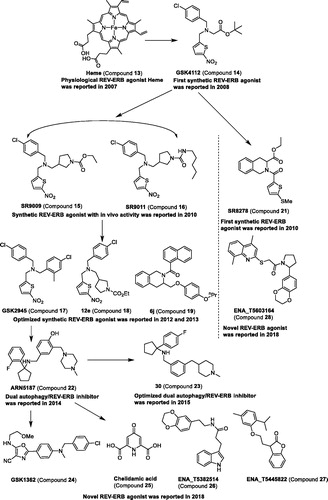
Table 2. Modulators targeting REV-ERBs.
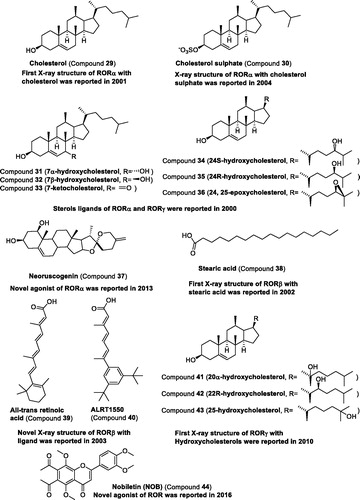
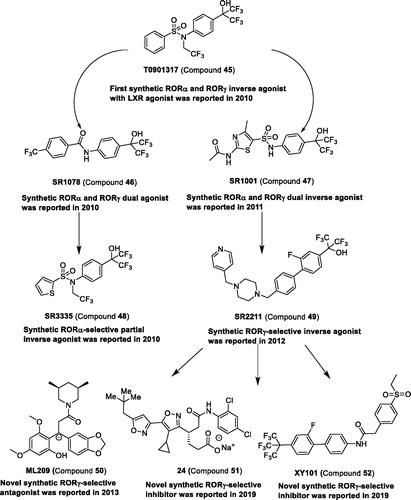
Table 3 Representative modulators targeting RORs.
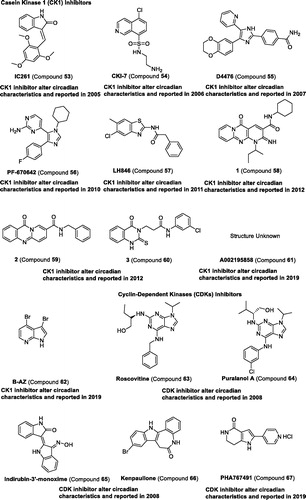
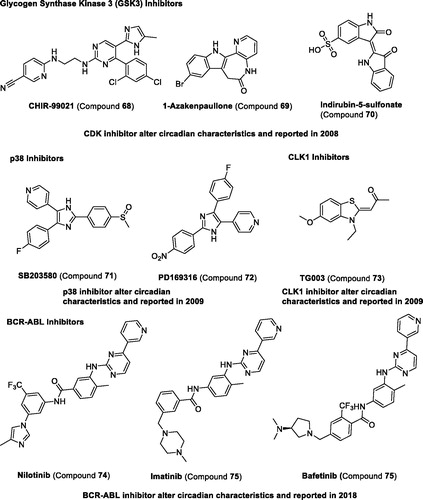
Table 4 Representative modulators targeting kinases.
Table 5. Representative modulators targeting epigenetic proteins.