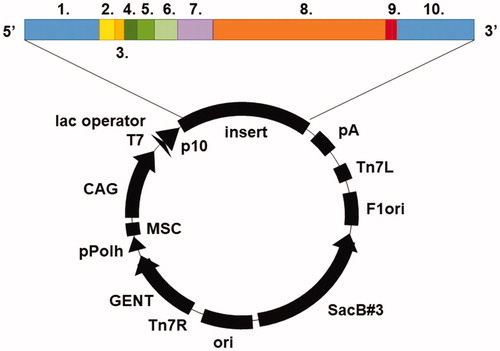
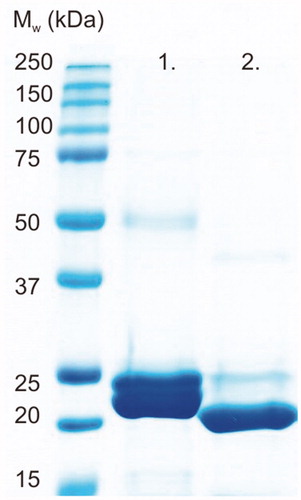
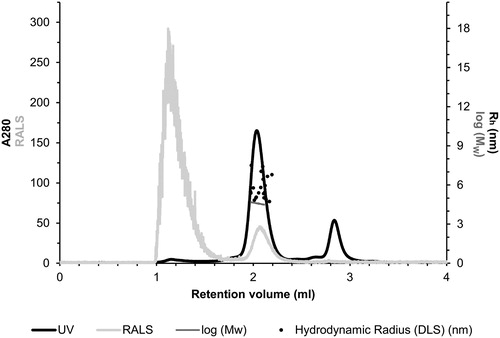
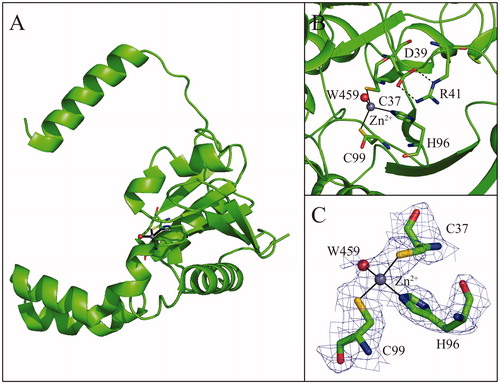
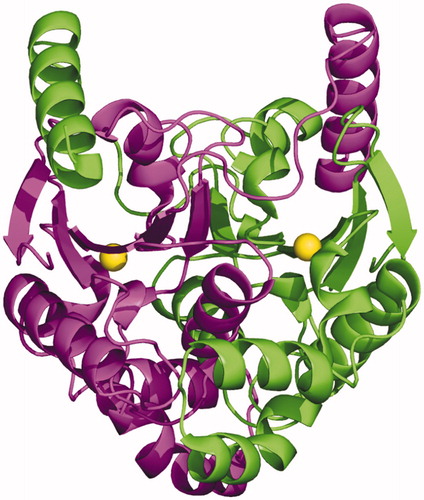
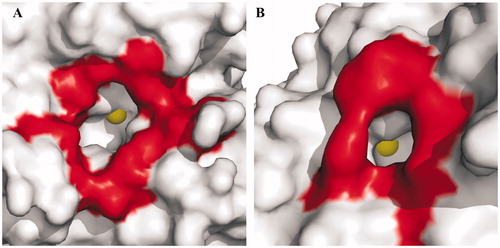
Figures & data
Table 1. Data collection and refinement statistics
Table 2. Kinetic data of TvaCA1. For comparison, kinetic parameters of hCA I, hCA II, and other representative β-CA enzymes are shown.
Table 3. β-CAs whose crystal structure has been determined