Figures & data
Figure 1. Chemical structure of the clinically used (palbociclib, abemaciclib and ribociclib) or currently in clinical trials (roniciclib) CDK inhibitors.
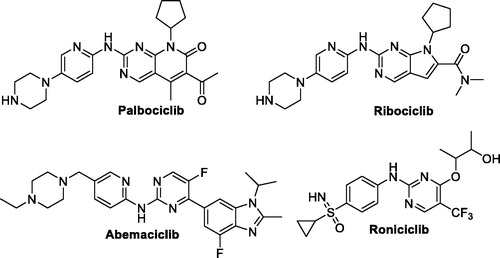
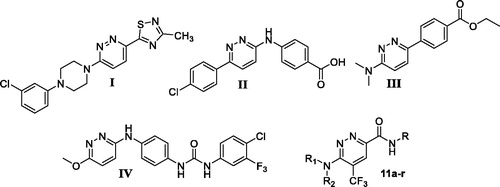
Figure 2. Chemical structure for some 3,6‐disubstituted pyridazine derivatives reported as efficient anticancer small molecules.
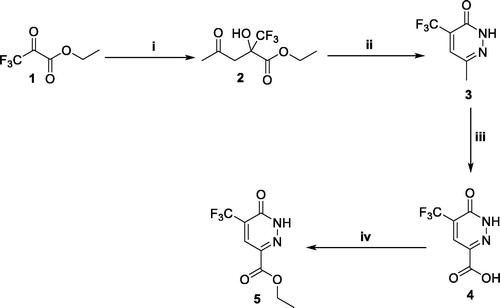
Scheme 1. Preparation of ester 5; reagents and conditions: (i) Acetone, DMF, L-proline, r.t., 48 h; (ii) NH2NH2, AcOH, reflux, 6 h; (iii) K2Cr2O7, Conc. H2SO4, 0 °C to r.t. overnight; (iv) EtOH, H2SO4, reflux, 4 h.
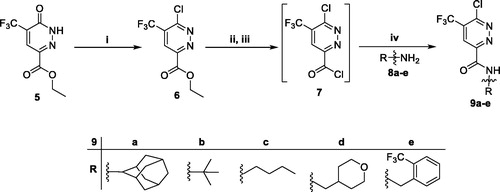
Scheme 2. Preparation of intermediateds 9a–e; reagents and conditions: (i) POCl3, 100 °C, 5 h; (ii) LiOH, THF, H2O, r.t., 1 h; (iii) SOCl2, DMF (catalytic), 1,2-dichloroethane, reflux, 3 h; (iv) Methylene chloride, Et3N, r.t., 4 h.
Scheme 3. Preparation of target pyridazines 11a–r; reagents and conditions: (i) 1,4-Dioxane, Hünig’s base, reflux, 6 h; (ii) Morpholine, 1,4-dioxane, Hünig’s base, reflux, 12 h; (iii) Primary amines 8a–e, EtOH, piperdine, reflux, 6 h.
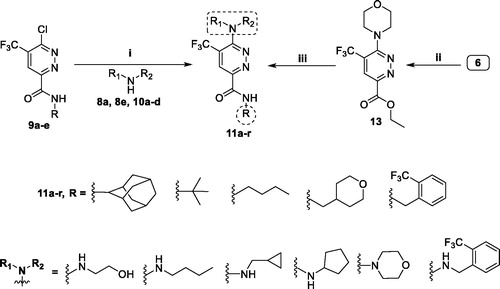
Table 1. Anti-proliferative activities for the newly prepared pyridazines (11a–r) towards T-47D, MDA-MB-231 and SKOV-3 cell lines.
Table 2. Cytotoxic impact for pyridazines 11l and 11m against non-tumorigenic MCF-10A human breast cell line, as well as mean tumour selectivity index (S. I.) (MCF-10A/T-47D and MDA-MB-231).
Figure 3. Influence of pyridazines 11l and 11 m on the distribution of cell cycle phases in breast cancer T-47D (A) and MDA-MB-231 (B) cells. Asterisk indicates a significant difference from control at p < 0.05.
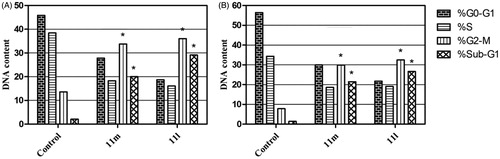
Figure 4. Percentage of apoptotic and necrotic cells in T-47D (A) and MDA-MB-231 (B), with respect to untreated control cells, upon treatment with pyridazines 11l and 11m.
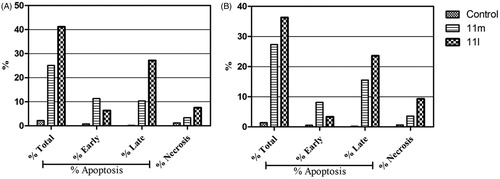
Figure 5. Influence of pyridazines 11l and 11m on the percentage of Annexin V+ apoptotic cells in T-47D cells.
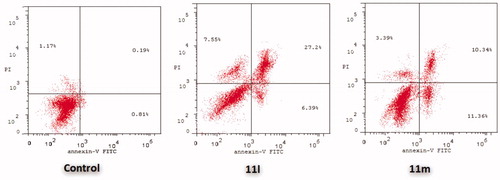
Figure 6. Influence of pyridazines 11l and 11m on the percentage of Annexin V+ apoptotic cells in MDA-MB-231 cells.
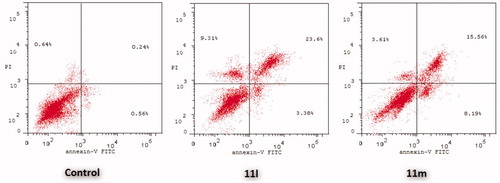
Figure 7. (A) 2D, and (B) 3D diagram for pyridazine 11e demonstrating its interactions within the CDK2 active site.
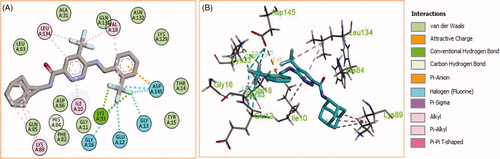
Figure 8. (A) 2D, and (B) 3D diagram for pyridazine 11h demonstrating its interactions within the CDK2 active site.
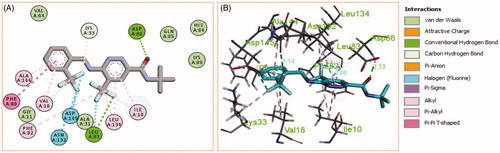
Figure 9. (A) 2D, and (B) 3D diagram for pyridazine 11l demonstrating its interactions within the CDK2 active site.
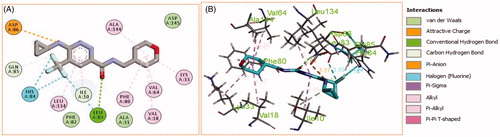
Figure 10. (A) 2D, and (B) 3D diagram for pyridazine 11m demonstrating its interactions within the CDK2 active site.
Table 3. Docking energy scores (S) for pyridazines 11a–r and the co-crystallized ligand Roniciclib (in in kcal/mol).
Table 4. The different bonding types and their distances (in Å) for pyridazines 11e, 11h, 11l, and 11m within the CDK2 active site.
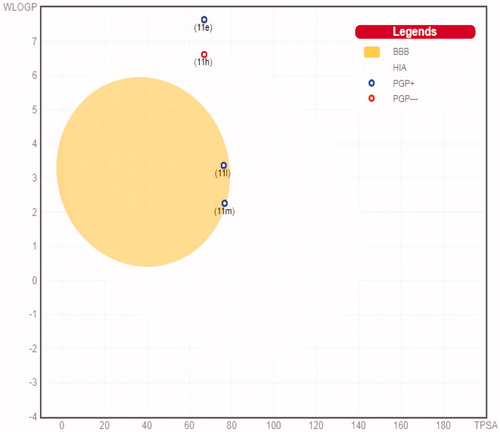
Table 5. IC50 values for the CDK2 inhibitory action of pyridazines 11e, 11h, 11l, and 11m.
Figure 12. The oral bioavailability radar chart for pyridazines 11e (A), 11h (B), 11l (C) and 11m (D), produced by swissADME online web tool.
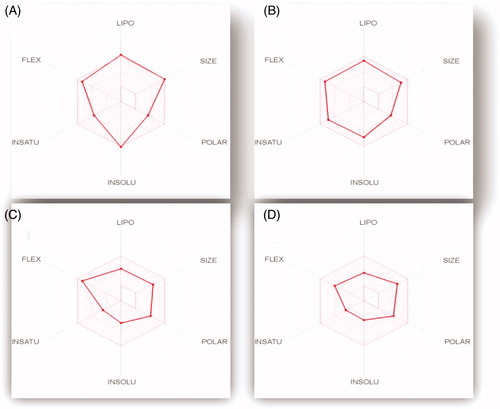
Table 6. The in silico predicted ADME for pyridazines 11e, 11h, 11l, and 11m.
Table 7. Compliance with Druglikeness rules (Lipinski, Veber, Egan, Ghose and Muegge), as well as PAINS and Brenk filters for target pyridazines 11e, 11h, 11l, and 11m.