Figures & data
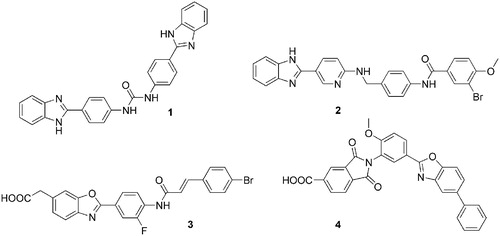
Figure 2. Workflow applied for similarity searches in eMolecules database. Literature inhibitors were first used as reference compounds, then highly potent newly identified inhibitors served as reference structures.
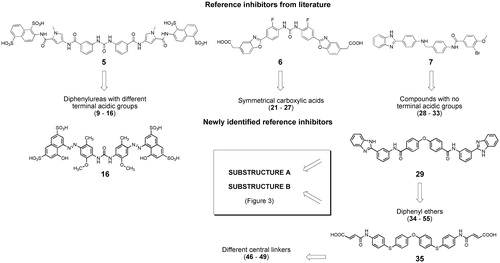
Figure 3. Workflow and query substructures used for virtual screening of heparanase inhibitors performed on SciFinder subset of commercially available compounds.
Table 1. Heparanase inhibitory activity (fondaparinux assay on human GS3 heparanase) of the most potent commercially available compounds selected through virtual screening.
Figure 4. Left: best docking pose obtained for sulphonyl-diphenyl urea 16 (green carbons). The docking pose of the reference inhibitor 6 (orange carbons) is shown for comparison. Right: docking pose obtained for terephthalamide derivative 61.
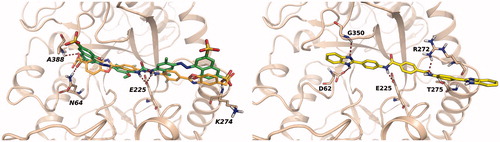
Figure 5. Left: overall superposition of docking poses obtained for compounds 21 (orange), 24 (green), 34 (light blue), 36 (yellow), 48 (gray), 49 (purple), 50 (pink), 57 (cyan), 58 (magenta) and 59 (white) selected through cluster analysis performed on structural interaction fingerprints. Right: superposition of best docking poses obtained for compounds 21 (orange carbons) and 24 (green carbons).
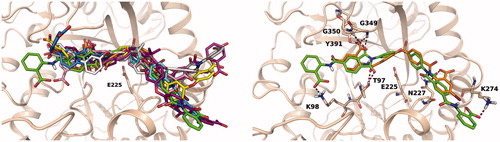
Table 2. Inhibition of invasive activity of HT1080, U87MG, and U2OS human cancer cell lines by newly discovered heparanase inhibitors.a
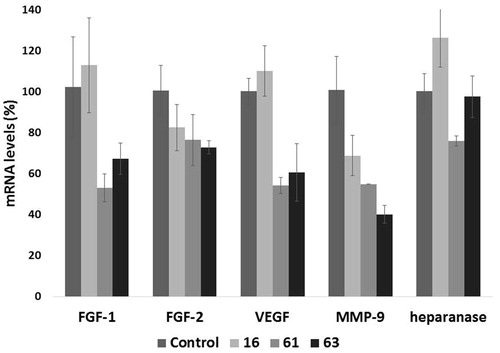
Figure 6. Effect of compounds 16, 61 and 63 (1 µM) on the expression of select angiogenesis-related genes. The expression levels of FGF-1, FGF-2, VEGF, MMP-9, and heparanase mRNA in HT1080 cells, upon 24 h of treatment, were measured through real-time qPCR analysis. Results are expressed as % with respect to untreated cells.