Figures & data
Table 1. Activity for TAM kinases of methoxy phenyl pyrrolopyrimidines.
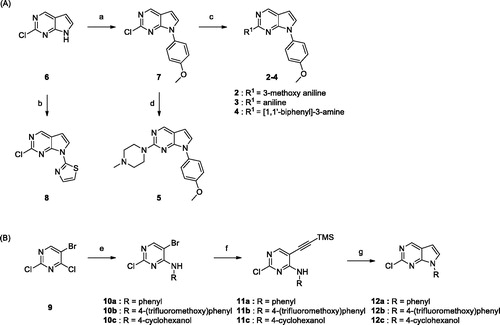
Scheme 1. Reagents and conditions: (a) (OH)2B-Ar, Cu(OAc)2, pyridine, 4 Å MS, CH2Cl2, rt, 6–24 h; (b) 2-bromothiazole, CuI, K3PO4, 1,2-trans-cyclohexanediamine, THF, 110 °C, 24 h; (c) BINAP, Pd2(dba)3, Cs2CO3, dioxane, 100 °C, 8 h; (d) HCl, i-PrOH, MW 160 °C, 1 h; (e) 4-(trifluoromethoxy)aniline, TEA, i-PrOH; (f) ethynyltrimethylsilane, Pd(PPh3)2, CuI, TEA, toluene, 80 °C, 4 h; (g) TBAF, THF, 60 °C, 4 h.
Scheme 2. Reagents and conditions: (a) amines, Pd2(dba)3, BINAP, Toluene, Cs2CO3, 100 °C, 6 − 24 h; (b) H2, Pd/C, MeOH, rt, 3 − 9 h; (c) 7, 8, or 12, HCl, i-PrOH, MW 160 °C, 1 h; (d) DIBAL, toluene, 0 °C, 3 h; (e) 1-methylpiperazine, NaBH(OAc)3, AcOH, DCE, rt, 12 h.
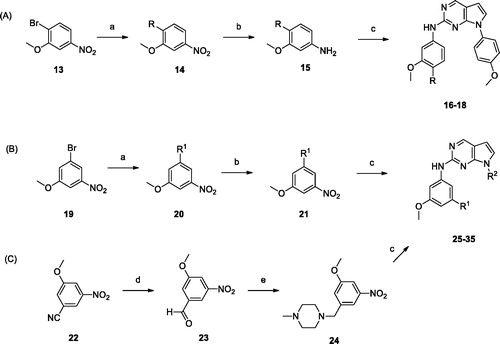
Figure 2. Predicted docking orientation of 16 and 25 with the Mer kinase domain (PDB ID: 3TCP). Docking mode of (a) 16 and (b) 25 with Mer. 2 D-interaction diagram of the binding model of (c) 16 and (d) 25. Estimated binding energies were −7.52 kcal/mol and −8.26 kcal/mol for 16 and 25, respectively. Hydrogen bonds and a salt bridge between the ligand and the backbone are shown in dashed lines. The docking study was performed by AutoDock Vina.
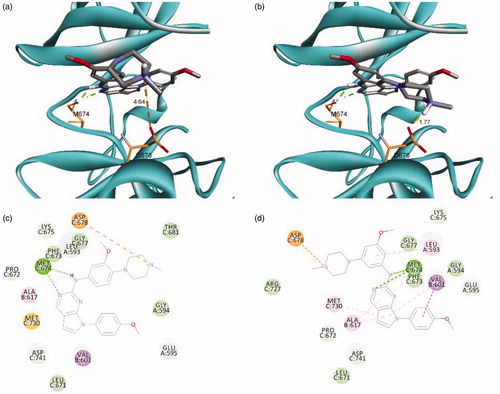
Table 2. Activities of 3-methoxy aniline derivatives of compound 3.
Table 3. Inhibitory activity of R4 derivatives.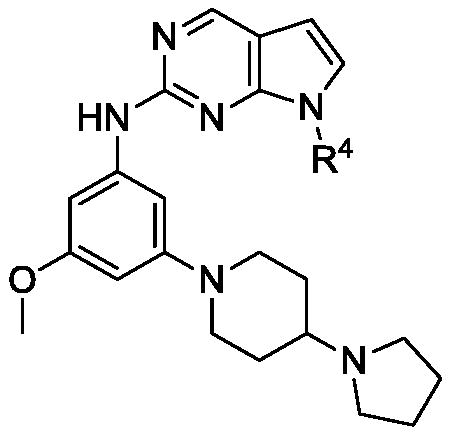
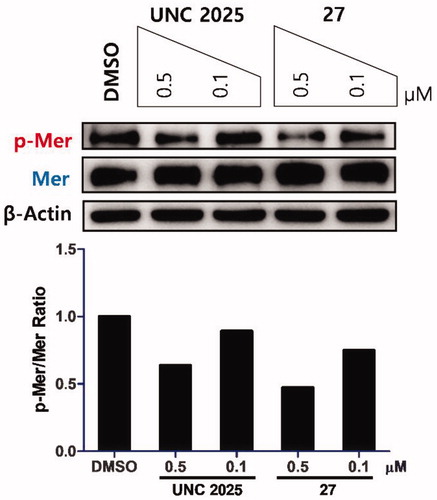
Figure 3. Inhibitory effect of 27 on Mer phosphorylation in a Mer-overexpressed human gastric cancer cell line, MKN28. Cells were treated with the indicated compounds at 0.5 and 0.1 μM for 1.5 h. UNC2025 and β-actin were used as a positive control and a loading control, respectively. Western blot analysis for phosphorylated and total Mer from a representative experiment is shown. Bar graph represents the relative intensities of the total and phosphorylated Mer as determined by band densitometry using image analysis software.