Figures & data
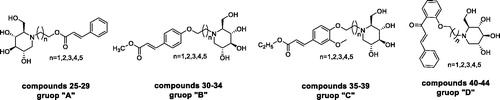
Scheme 1. Synthesis of intermediate products and target products of N-alkyl-deoxynojirimycin derivatives. Reagents and condition: (a) K2CO3, acetone, dibromo alkane, 65 °C, overnight or Et3N, acetone, dibromo alkane, 65 °C; (b) K2CO3, DMF, 85 °C, 6 h.
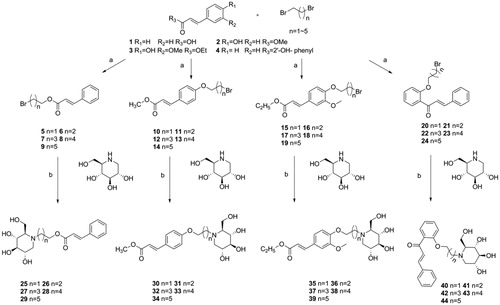
Table 1. In vitro α-glucosidase inhibitory activity of compound 25–44.
Figure 2. Kinetic analysis of α-glucosidase inhibition by compounds 43, 40, and 34. (A) The Lineweaver–Burk plots in the absence and presence of different concentrations of compound 43; (B) The Lineweaver–Burk plots in the absence and presence of different concentrations of compound 40; (C) The Lineweaver–Burk plots in the absence and presence of different concentrations of compound 34.
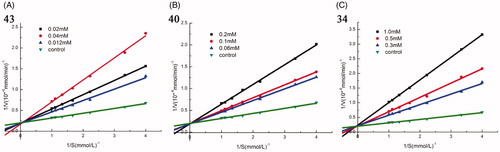
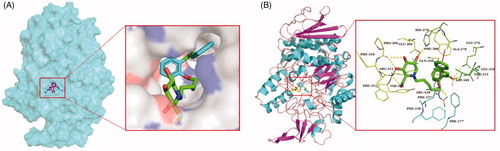
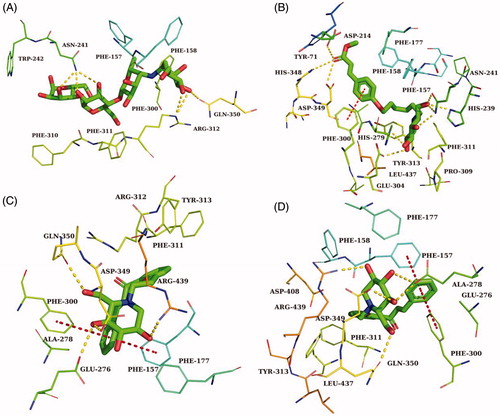
Table 2. The detailed information of molecular docking results of compounds 34, 40, 41, 43, and acarbose.