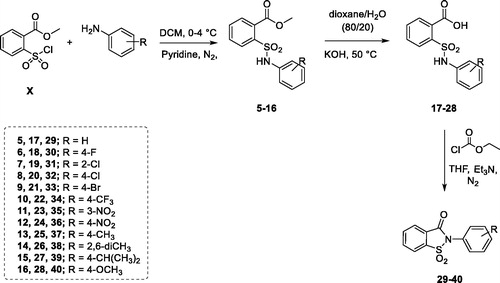
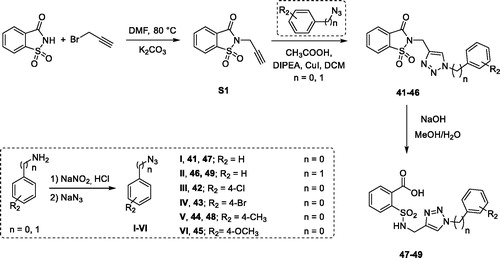
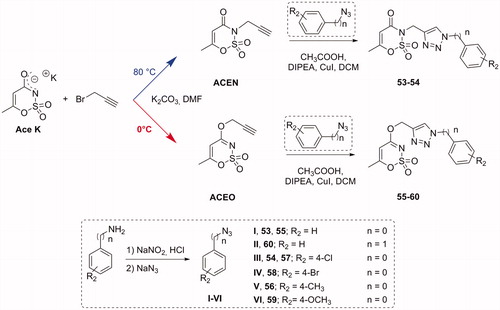
Figures & data
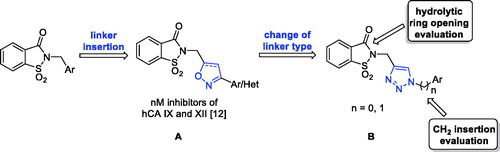
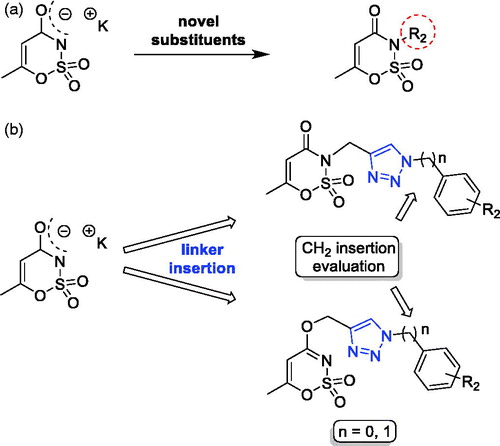
Figure 1. (a) The first design strategy proposed in this paper; (b) design and retrosynthetic approach for novel hCAs potential inhibitors.
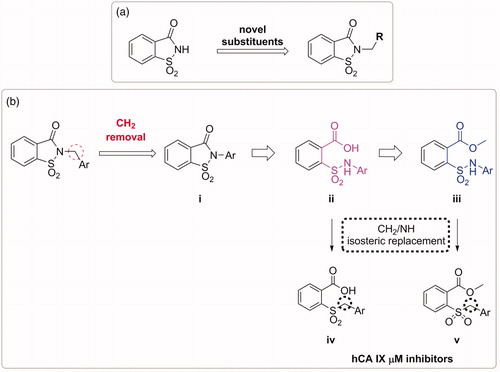
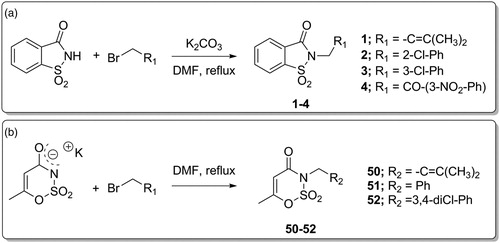
Table 1. Inhibition data of selected human CA isoforms (hCA I, II, IX and XII) with saccharin-based derivatives 1–49 reported here and the standard sulphonamide inhibitor acetazolamide (AAZ) by a stopped flow CO2 hydrase assayCitation49.
Table 2. Inhibition data of selected human CA isoforms (hCA I, II, IX and XII) with acesulfame-based derivatives 50–60 reported here and the standard sulphonamide inhibitor acetazolamide (AAZ) by a stopped flow CO2 hydrase assayCitation49.
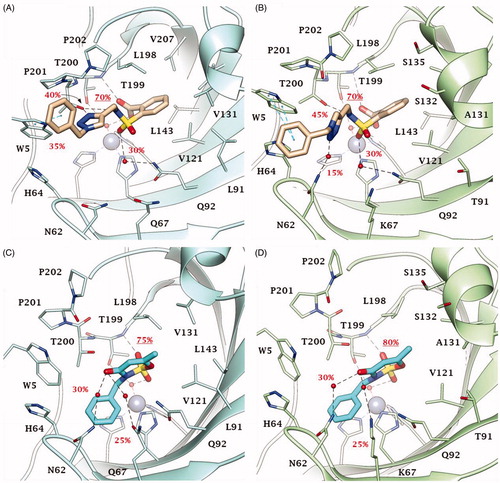
Figure 4. Predicted binding mode of compounds 2 and 46 into (A, C) CA IX and (B, D) CA XII active site. H-bonds and π-π stackings are represented as black and blue dashed lines, respectively. Dashed bonds occupancy over the MD simulation is indicated as percentage, among which underlined is the occupancy of the anchorage to the zinc-bound water. Water molecules are shown as red spheres. Amino acids are labelled with one letter symbols: A, Ala; H, His; K, Lys; L, Leu; N, Asn; P, Pro; Q, Gln; S, Ser; T, Thr; V, Val; W, Trp.
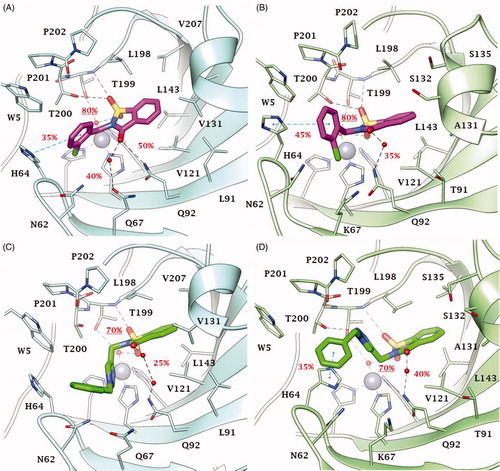
Figure 5. Predicted binding mode of compounds 49 and 51 into (A, C) CA IX and (B, D) CA XII active site. H-bonds and π-π stackings are represented as black and blue dashed lines, respectively. Dashed bonds occupancy over the MD simulation is indicated as percentage, among which underlined is the occupancy of the anchorage to the zinc-bound water. Water molecules are shown as red spheres. Amino acids are labelled with one letter symbols: A, Ala; H, His; K, Lys; L, Leu; N, Asn; P, Pro; Q, Gln; S, Ser; T, Thr; V, Val; W, Trp.