Figures & data
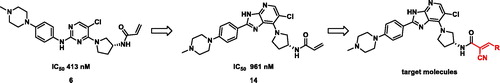
Scheme 1. Reagents and conditions: (a) (R)-3-Boc-aminopyrrolidine, K2CO3, MeCN, RT, 17 h, 53%; (b) Fe powder, AcOH, 40 °C, 1 h, 66%; (c) 4–(4-Methylpiperazin-1-yl)benzaldehyde, FeCl3, DMF, 120 °C, 16 h, 32%; (d) TFA, DCM, RT, 1 h, 70%; (e) Cyanoacetic acid, EDCI, HOBt, DIPEA, DMF, RT, 16 h, 56%; (f) Aldehyde, piperidine, 2-propanol, 60 °C, 2 h, 20%; (g) Na2CO3, aqueous THF, acryloyl chloride, 0 °C, 2 h, 47%.
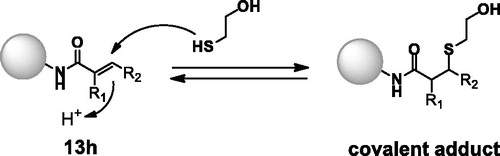
Figure 3. Predicted binding mode of 13h with C174 of TAK1 kinase domain (PDB: 4L52)Citation25. Imidazopyridine core interacts with hinge region of TAK1 and pyridinyl nitrogen forms a hydrogen bond with D175. The estimated free energy of binding was found to be −9.65 kcal/mol and −7.05 kcal/mol for 13h and 14, respectively. Covalent docking study was performed using Autodock via flexible side chain methodCitation26. The figure was visualised using Discovery Studio 2020 Visualiser.
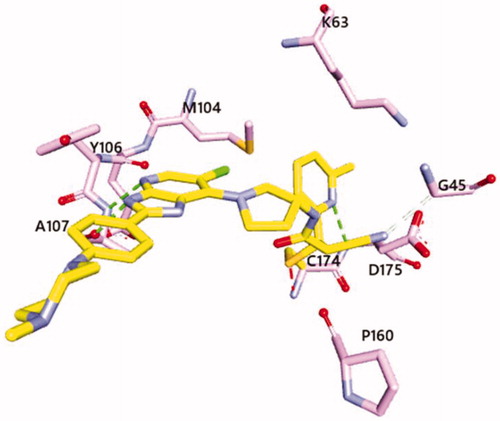
Table 1. TAK1 enzymatic assay with imidazopyridine derivatives.
Table 2. Kinase profile of 13h (1 µM).
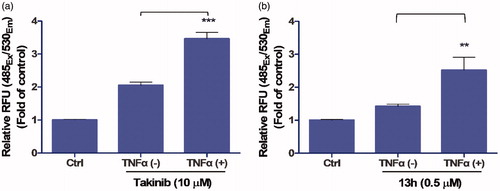
Figure 4. Effect of 13h on MDA-MB-231 cells. Caspase-3/7 activity of (a) Takinib (10 µM) and (b) 13h (0.5 µM) in the presence or absence of TNFα. Data are reported as the mean ± SD (n = 3). *** p < 0.001, ** p < 0.01.