Figures & data
Scheme 1. Synthetic route for the preparation of compounds 4a-4c and precursors. Reagents and conditions: (i) microwave, DMF, stirred at 100 W for 1 h; (ii) acetonitrile, pyridine, stirred for 1–2 h, room temperature; (iii) oxalyl chloride, stirred for 30 min, room temperature.
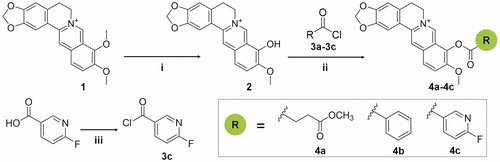
Table 1. Inhibitory activities and binding affinities of the compounds towards different GH18 chitinases.
Table 2. Details of data collection and structure refinement.
Figure 1. Crystal structure of SmChiB in complex with berberine. (A) Structure of berberine. The conjugate tetracycle plane is shown in green-cyan, the 9-O-methoxy and 10-O-methoxy moieties are shown in pink. (B) Binding mode of berberine in the active pocket of SmChiB. Berberine is shown in stick representation with carbon atoms in green. The aromatic residues that stack with berberine are labelled and shown in stick representation with carbon atoms in yellow. The amino residues forming the hydrophobic cavity extended near the +2 subsite are labelled and shown as stick representation with carbon atoms in blue. The numbers indicate the subsite to which the berberine is bound.
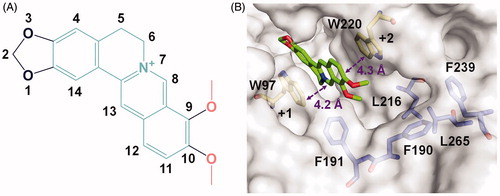
Figure 2. Modelled complex structures of compounds 4a-4c in SmChiB. Details of the interaction of compound 4a (A), 4 b (B) and 4c (C) with SmChiB. Compound 4a is shown in blue, compound 4 b is shown in magenta and compound 4c is shown in cyan. Residues that participate in binding are shown in yellow. Hydrogen bonds are shown as dashed black lines. The numbers indicate the subsite to which the compounds are bound.
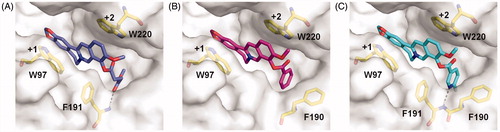
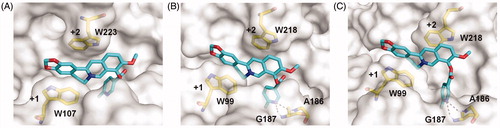
Figure 3. Modelled structures of compound 4c in OfChtI (A), HsCht (B), and hAMCase (C). Compound 4c is shown in stick representation with carbon atoms in cyan. Amino acids that interact with compound 4c are labelled and shown in stick representation with carbon atoms in yellow. Hydrogen bonds are shown as dashed black lines. The numbers indicate the subsite to which compound 4c is bound.