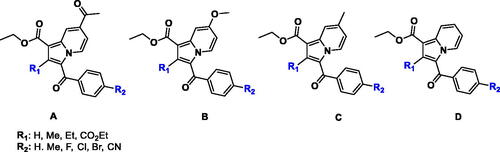
Figures & data
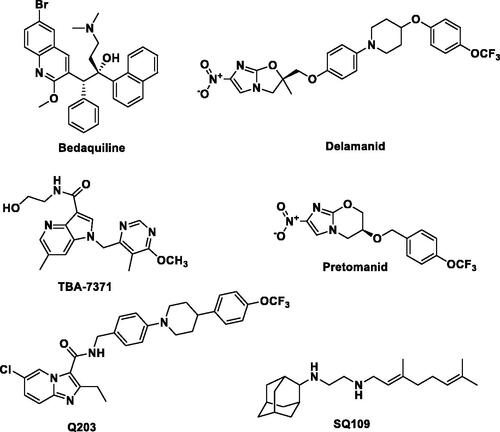
Figure 1. Chemical structures of approved anti-TB drugs (Bedaquiline, Delamanid and Pretomanid) and anti-TB compounds undergoing clinical trial (Q203, TBA-7371 and SQ109).
Scheme 1. Synthesis of 1,2,3-trisubstiuted indolizine derivatives (2a–2f, 3a–3d, 4a–4c): Reagents and conditions (i) pyridine, dry acetone, stir at room temperature, 5 h; (ii) ethyl propialate/ethy1 2-butynoate/diethy1 2- butynedioate water, stir 80 °C, 3 h
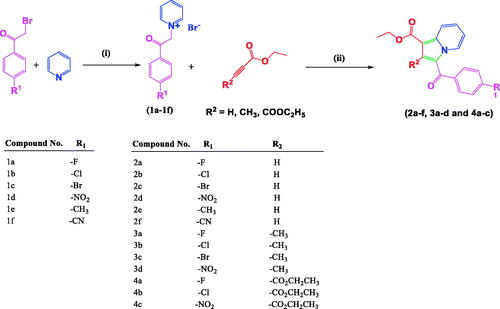
Figure 3. The crystal structure of diethyl 3-(4-chlorobenzoyl)indolizine-1,2-dicarboxylate (4b). ORTEP drawn with 50% ellipsoidal probability. Intra-molecular C–H···O·H bonds are shown as dotted lines.
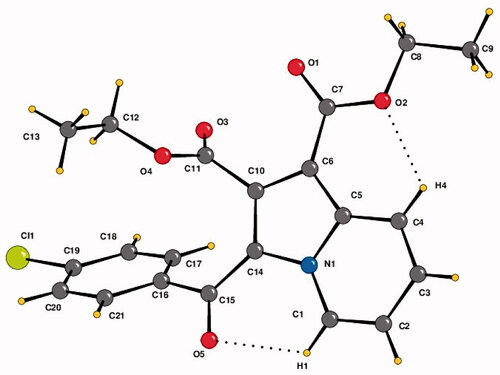
Figure 4. Crystal packing depicting C–H···O·H-bonds, π–π stacking interaction and O1 = C7···C5–N1 short contact in the compound 4b. Cg1 is the centre of gravity of N1/C1–C5 ring. The dotted lines represent intermolecular interactions. Non-interacting hydrogens have been omitted for clarity.
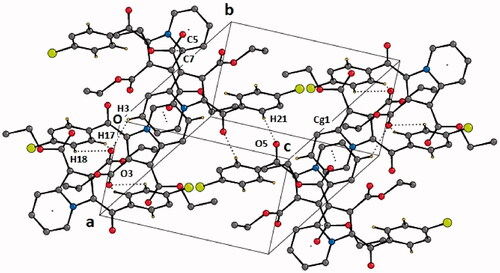
Table 1. List of intramolecular and intermolecular interactions in the crystal structure of diethyl 3–(4-chlorobenzoyl)indolizine-1,2-dicarboxylate (4b).
Figure 5. Hirshfeld surfaces mapped with (a) dnorm (b) de (c) shape-index, (d) curvedness and (e) fragment patches for the compound 4b.
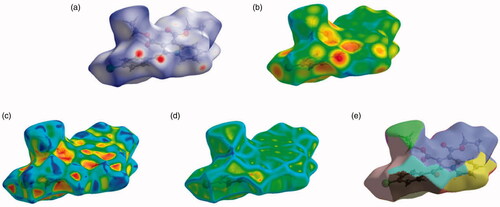
Figure 6. Two-dimensional fingerprint plot for the compound 4b showing the contributions of individual types of interactions: (a) all intermolecular contacts, (b) H···H contacts, (c) O···H/H···O contacts, (d) C···H/H···C contacts, (e) Cl···H/H···Cl contacts, (f) C···C contacts, (g) C···O/O···C contacts, (h) N···C/C···N. The outline of the full fingerprint is indicating in grey. The related surface patches associated with the specific contacts with the dnorm mapped are illustrated by surfaces to the left.
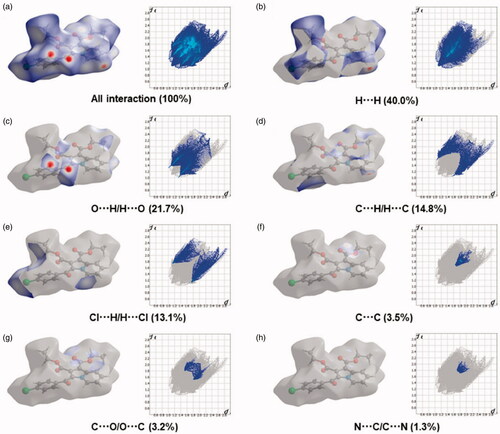
Table 2. In vitro anti-mycobacterial activity of 1,2,3-trisubstituted indolizine derivatives (2a–2f, 3a–3d, and 4a–4c) against H37Rv and MDR strains of Mycobacterium tuberculosis. 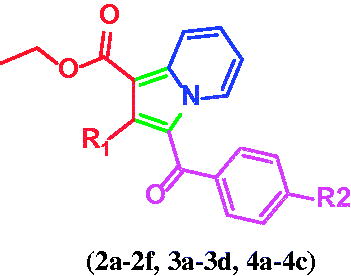
Table 3. Calculated ADMET descriptors and toxicity parameters of indolizine derivatives (2a–2f, 3a–3d, and 4a–4c).
Figure 7. The two reported “in and out” conformations of Tyr158 in mycobacterial InhA enzyme. PDB ID for the in conformation is 5G0S, and that for the out conformation is 5G0U.
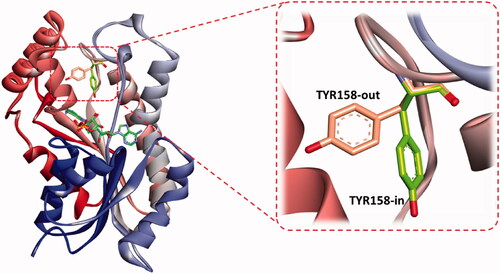
Figure 8. Comparative binding mode of indolizine 2d into InhA binding domain with (a) in-Tyr 158 (PDB ID: 5G0S) and (b) out-Tyr 158 conformations (PDB ID: 5G0U), respectively. The ligand, NAD and the receptor were represented as sticks in solmon, yellow and cyan colours respectively. The hydrogen bonds are shown as green dotted lines.
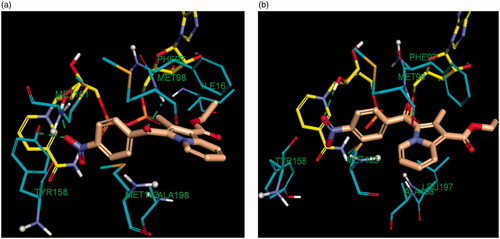
Figure 9. Predicted binding interaction of indolizines (2a, 2b, 2d, 3a, 3b, 3d, 4a–4c) with InhA binding domain (PDB 5G0S). Ligand and receptor were represented as solmon and cyan respectively. Hydrogen bonding contact is shown with green dotted lines. π–π, π–sulphur and hydrophobic interactions are shown with magenta, gold and violet, respectively. For displaying key bingind interaction, NAD has been omitted.
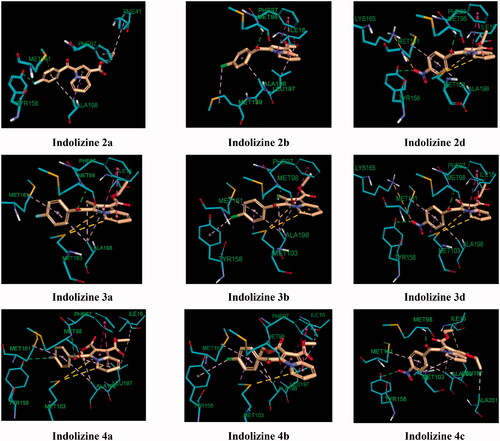
Table 4. Docking results of the indolizine derivatives 2a–2f, 3a–3d and 4a–4c into the InhA binding domain (PDB 5G0S).
Figure 10. Predicted binding interaction of indolizines (4a, 4c) with InhA binding domain (PDB 5G0S). Ligand, NAD and receptor were represented as solmon, yellow and cyan respectively. Hydrogen bonding contact is shown with green dotted lines.
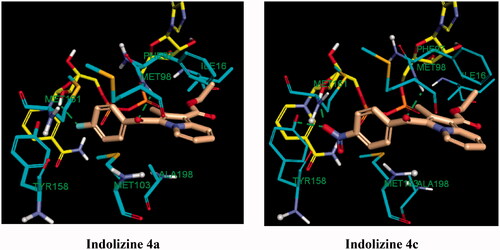
Figure 11. Predicted binding interaction of indolizines (4a–4d) with anthranilate phosphoribosyl binding domain (PDB 3R6C). Ligand and receptor were represented as solmon and cyan respectively. Hydrogen bonding contact is shown with green dotted lines. Halogen bonding and hydrophobic intreactions are shown with cyan and violet respectively.
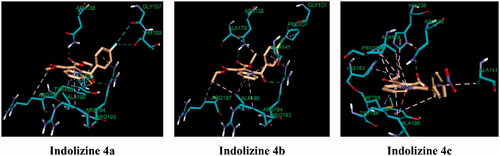
Table 5. Docking results of the indolizine derivatives 4a–4c into the anthranilate phosphoribosyl binding domain (PDB 3R6C).