Figures & data
Figure 2. Structure and functions of mTOR complex 1. The best-known targets of mTORC1 phosphorylation S6K1 and 4EBP1 have a vital function in protein synthesis. Phosphorylated S6K1 consequently phosphorylates ribosomal protein S6 and commences cap-independent translation. Activated 4EBP1 relieves its inhibitory function towards eIF4E and initiates cap-dependent translation. Ribosomal biogenesis is enhanced by stimulated translation of 5′TOP mRNA via 4EBP1 and LARP1. Pol I and Pol III are activated by phosphorylation of TIF1A and inhibition of Maf1, respectively. Autophagy-related ULK1/ATG13/FIP200 protein complex is inhibited by mTORC1 phosphorylation, as well as WIPI2 (positive regulator of autophagy). On the other hand, autophagy suppressor DAP1 is activated. Protein expression of lipid and cholesterol homeostasis-involved genes is managed by transcription factors SREBP and PPAR-γ. mTORC1-mediated phosphorylation of lipin-1 alleviates SREBP inhibition. Several glycolytic enzyme genes are indirectly modulated by mTORC1 via transcription factors HIF1α and Myc. Expression of HIF1α is regulated either at the level of translation or via inhibited GSK3/Foxk1 pathway.
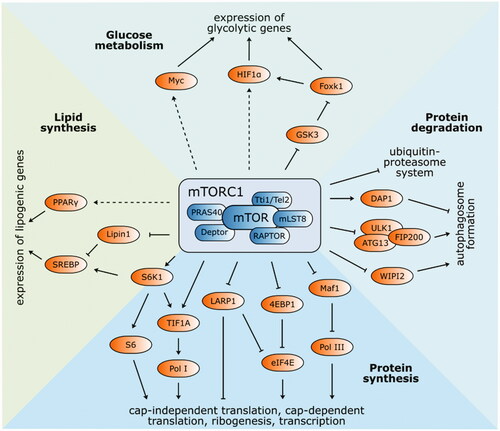
Figure 3. Upstream regulators of mTORC1 and mTORC2. Growth factors (insulin/IGF-1) bind to their receptors that phosphorylate IRS and activate PI3K. PI3K-IRS complex converts phosphatidylinositol-4,5-bisphosphate (PIP2) to phosphatidylinositol-3,4,5-trisphosphate (PIP3), which recruits phosphoinositide-dependent kinase 1 (PDK1) to activate Akt. PIP2-PIP3 conversion is counteracted by PTEN. Akt inhibits the TSC complex that acts as GTPase-activating (GAP) protein for Rheb. mTORC1 is activated by GTP-bound Rheb protein. Thus, when Akt activity is stimulated by PIP3, it phosphorylates TSC1/2 and switches off its inhibiting activity towards Rheb. Akt activates mTORC1 directly via phosphorylation of PRAS40. Amino acids induce activation of Rag proteins, which mediates translocation of mTORC1 to lysosomal surface. The intra lysosomal amino acids activate mTORC1 in an arginine-dependent manner via interaction of transporter SLC38A9 with the Rag-Ragulator-v-ATPase complex. Cytosolic amino acids engage the negative regulator GATOR1 (tethered to the lysosome by KICSTOR) and GATOR2. A direct leucine sensor Sestrin2 and arginine sensor CASTOR1 dissociate from GATOR2 in the presence of amino acids and release its inhibitory effect on GATOR1, thus positively regulate the mTORC1 pathway. AMPK negatively regulates mTORC1 either directly by Raptor phosphorylation, or via TSC1/2-Rheb axis. mTORC2 activation is PI3K-sensitive. Regulators of the mTOR pathway are depicted in green (positive) and red (negative). Dashed lines indicate indirect regulation.
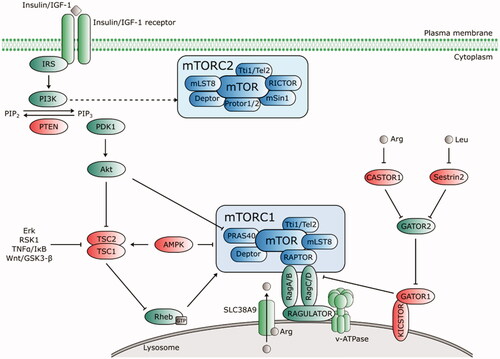
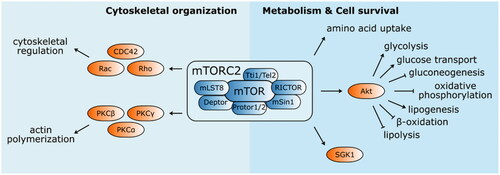
Figure 4. Structure and functions of mTOR complex 2. mTORC2 mainly phosphorylate several kinases from AGC family (PKCα, PKCβ, PKCγ, PKCε, PKCη/Λ, PKCδ, PKCθ, and SGK1). It is involved in the cytoskeletal organisation by modulating actin polymerisation and cytoskeletal structure regulation via PKCα and Rho family of small GTPases (Rac, Rho, and CDC42), respectively. mTORC2 also activates Akt, an important enhancer and suppressor of several metabolic pathways engaging mTORC2 in metabolism regulation and cell survival.